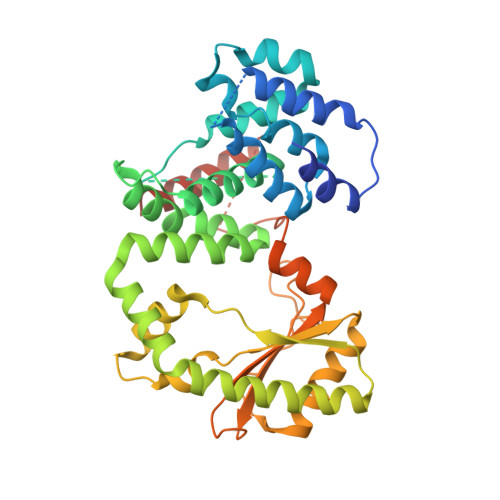
Crystallographic and solution structure of the N-terminal domain of the Rel protein from Mycobacterium tuberculosis
Singal, B., Balakrishna, A.M., Nartey, W., Manimekalai, M.S.S., Jeyakanthan, J., Gruber, G.(2017) FEBS Lett 591: 2323-2337
- PubMed: 28672070
- DOI: https://doi.org/10.1002/1873-3468.12739
- Primary Citation of Related Structures:
5XNX - PubMed Abstract:
Modulation of intracellular guanosine 3',5'-bispyrophosphate ((p)ppGpp) level, the effector of the stringent response, is crucial for survival as well as optimal growth of prokaryotes and, thus, for bacterial pathogenesis and dormancy. In Mycobacterium tuberculosis (Mtb), (p)ppGpp synthesis and degradation are carried out by the bifunctional enzyme MtRel, which consists of 738 residues, including an N-terminal hydrolase- and synthetase-domain (N-terminal domain or NTD) and a C-terminus with a ribosome-binding site. Here, we present the first crystallographic structure of the enzymatically active MtRel NTD determined at 3.7 Å resolution. The structure provides insights into the residues of MtRel NTD responsible for nucleotide binding. Small-angle X-ray scattering experiments were performed to investigate the dimeric state of the MtRel NTD and possible substrate-dependent structural alterations.
Organizational Affiliation:
School of Biological Sciences, Nanyang Technological University, Singapore, Singapore.