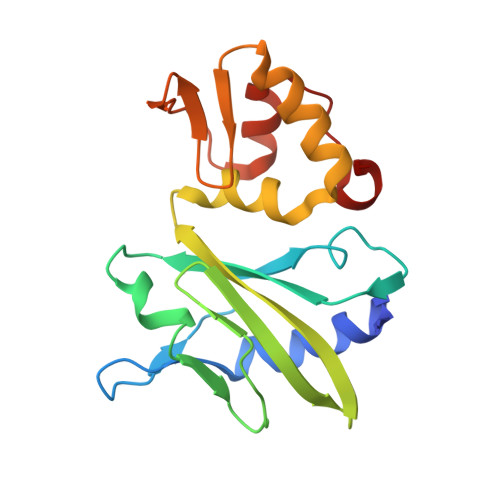
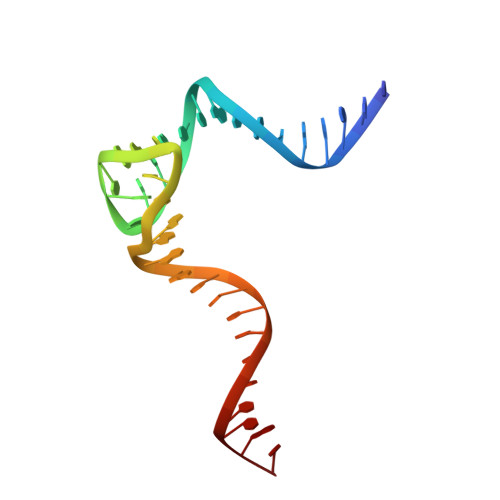
Crystal structure of an RNA-cleaving DNAzyme.
Liu, H., Yu, X., Chen, Y., Zhang, J., Wu, B., Zheng, L., Haruehanroengra, P., Wang, R., Li, S., Lin, J., Li, J., Sheng, J., Huang, Z., Ma, J., Gan, J.(2017) Nat Commun 8: 2006-2006
- PubMed: 29222499
- DOI: https://doi.org/10.1038/s41467-017-02203-x
- Primary Citation of Related Structures:
5XM8, 5XM9, 5XMA - PubMed Abstract:
In addition to storage of genetic information, DNA can also catalyze various reactions. RNA-cleaving DNAzymes are the catalytic DNAs discovered the earliest, and they can cleave RNAs in a sequence-specific manner. Owing to their great potential in medical therapeutics, virus control, and gene silencing for disease treatments, RNA-cleaving DNAzymes have been extensively studied; however, the mechanistic understandings of their substrate recognition and catalysis remain elusive. Here, we report three catalytic form 8-17 DNAzyme crystal structures. 8-17 DNAzyme adopts a V-shape fold, and the Pb 2+ cofactor is bound at the pre-organized pocket. The structures with Pb 2+ and the modification at the cleavage site captured the pre-catalytic state of the RNA cleavage reaction, illustrating the unexpected Pb 2+ -accelerated catalysis, intrinsic tertiary interactions, and molecular kink at the active site. Our studies reveal that DNA is capable of forming a compacted structure and that the functionality-limited bio-polymer can have a novel solution for a functional need in catalysis.
Organizational Affiliation:
State Key Laboratory of Genetic Engineering, Collaborative Innovation Center of Genetics and Development, Department of Physiology and Biophysics, School of Life Sciences, Fudan University, Shanghai, 200438, China.