Selective oxidation of aliphatic C-H bonds in alkylphenols by a chemomimetic biocatalytic system
Du, L., Dong, S., Zhang, X., Jiang, C., Chen, J., Yao, L., Wang, X., Wan, X., Liu, X., Wang, X., Huang, S., Cui, Q., Feng, Y., Liu, S.J., Li, S.(2017) Proc Natl Acad Sci U S A 114: E5129-E5137
- PubMed: 28607077
- DOI: https://doi.org/10.1073/pnas.1702317114
- Primary Citation of Related Structures:
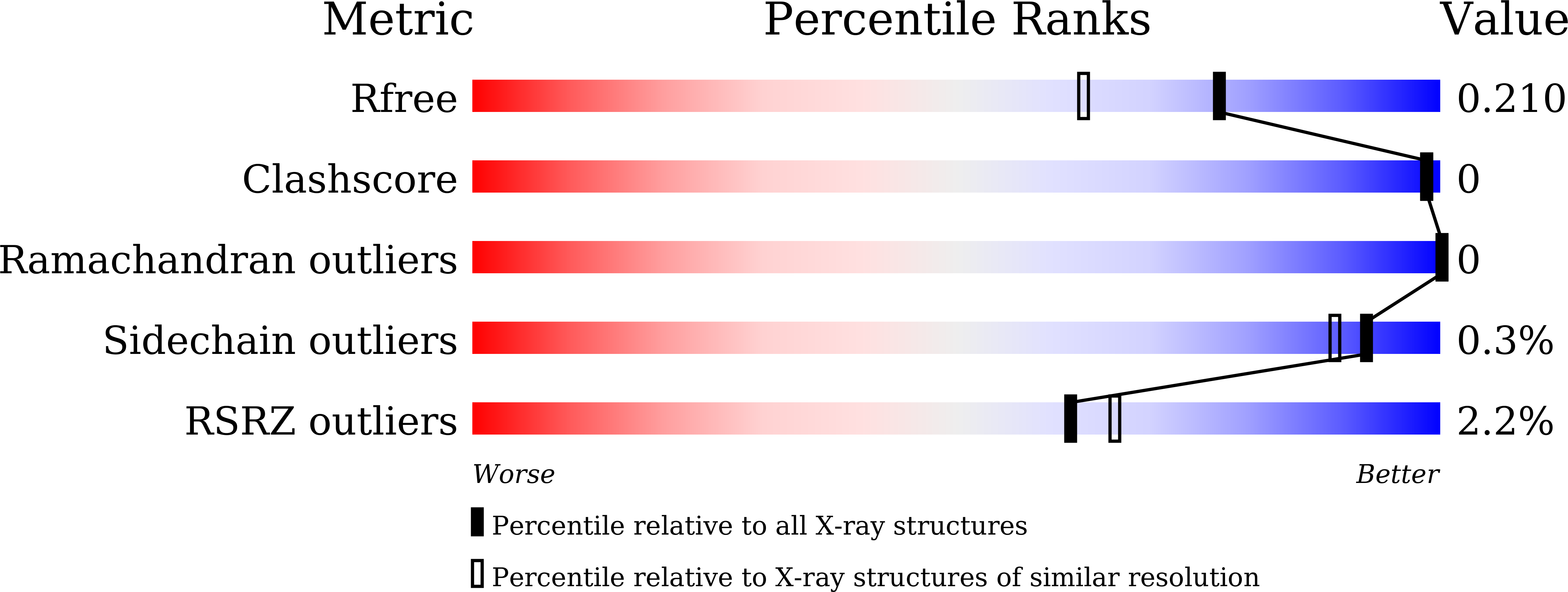
5GWE, 5XJN - PubMed Abstract:
Selective oxidation of aliphatic C-H bonds in alkylphenols serves significant roles not only in generation of functionalized intermediates that can be used to synthesize diverse downstream chemical products, but also in biological degradation of these environmentally hazardous compounds. Chemo-, regio-, and stereoselectivity; controllability; and environmental impact represent the major challenges for chemical oxidation of alkylphenols. Here, we report the development of a unique chemomimetic biocatalytic system originated from the Gram-positive bacterium Corynebacterium glutamicum The system consisting of CreHI (for installation of a phosphate directing/anchoring group), CreJEF/CreG/CreC (for oxidation of alkylphenols), and CreD (for directing/anchoring group offloading) is able to selectively oxidize the aliphatic C-H bonds of p - and m -alkylated phenols in a controllable manner. Moreover, the crystal structures of the central P450 biocatalyst CreJ in complex with two representative substrates provide significant structural insights into its substrate flexibility and reaction selectivity.
Organizational Affiliation:
Shandong Provincial Key Laboratory of Synthetic Biology, Qingdao Institute of Bioenergy and Bioprocess Technology, Chinese Academy of Sciences, Qingdao, Shandong 266101, China.