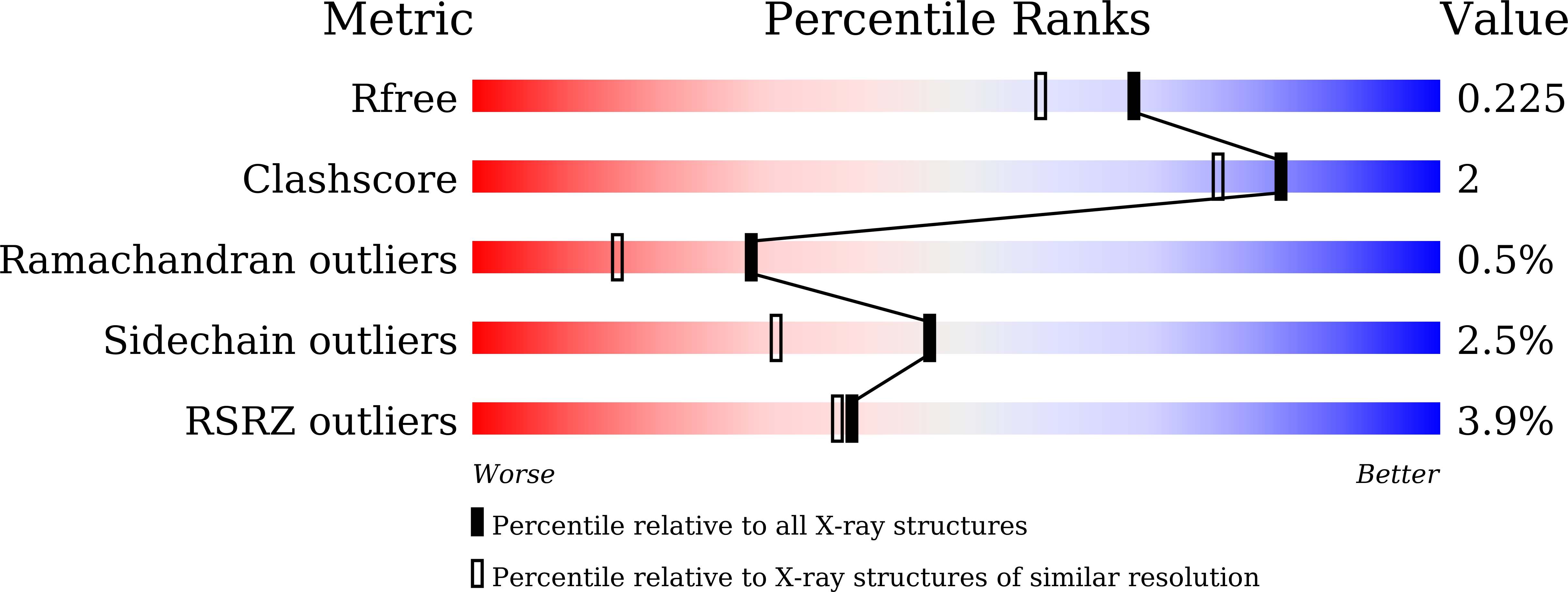
Crystal structure of a Ca2+-dependent regulator of flagellar motility reveals the open-closed structural transition
Shojima, T., Hou, F., Takahashi, Y., Matsumura, Y., Okai, M., Nakamura, A., Mizuno, K., Inaba, K., Kojima, M., Miyakawa, T., Tanokura, M.(2018) Sci Rep 8: 2014-2014
- PubMed: 29386625
- DOI: https://doi.org/10.1038/s41598-018-19898-7
- Primary Citation of Related Structures:
5X9A, 5YPX - PubMed Abstract:
Sperm chemotaxis toward a chemoattractant is very important for the success of fertilization. Calaxin, a member of the neuronal calcium sensor protein family, directly acts on outer-arm dynein and regulates specific flagellar movement during sperm chemotaxis of ascidian, Ciona intestinalis. Here, we present the crystal structures of calaxin both in the open and closed states upon Ca 2+ and Mg 2+ binding. The crystal structures revealed that three of the four EF-hands of a calaxin molecule bound Ca 2+ ions and that EF2 and EF3 played a critical role in the conformational transition between the open and closed states. The rotation of α7 and α8 helices induces a significant conformational change of a part of the α10 helix into the loop. The structural differences between the Ca 2+ - and Mg 2+ -bound forms indicates that EF3 in the closed state has a lower affinity for Mg 2+ , suggesting that calaxin tends to adopt the open state in Mg 2+ -bound form. SAXS data supports that Ca 2+ -binding causes the structural transition toward the closed state. The changes in the structural transition of the C-terminal domain may be required to bind outer-arm dynein. These results provide a novel mechanism for recognizing a target protein using a calcium sensor protein.
Organizational Affiliation:
Department of Applied Biological Chemistry, Graduate School of Agricultural and Life Sciences, The University of Tokyo, 1-1-1 Yayoi, Bunkyo-ku, Tokyo, 113-8657, Japan.