Design, synthesis, and evaluation of novel inhibitors for wild-type human serine racemase.
Takahara, S., Nakagawa, K., Uchiyama, T., Yoshida, T., Matsumoto, K., Kawasumi, Y., Mizuguchi, M., Obita, T., Watanabe, Y., Hayakawa, D., Gouda, H., Mori, H., Toyooka, N.(2017) Bioorg Med Chem Lett
- PubMed: 29277459
- DOI: https://doi.org/10.1016/j.bmcl.2017.12.021
- Primary Citation of Related Structures:
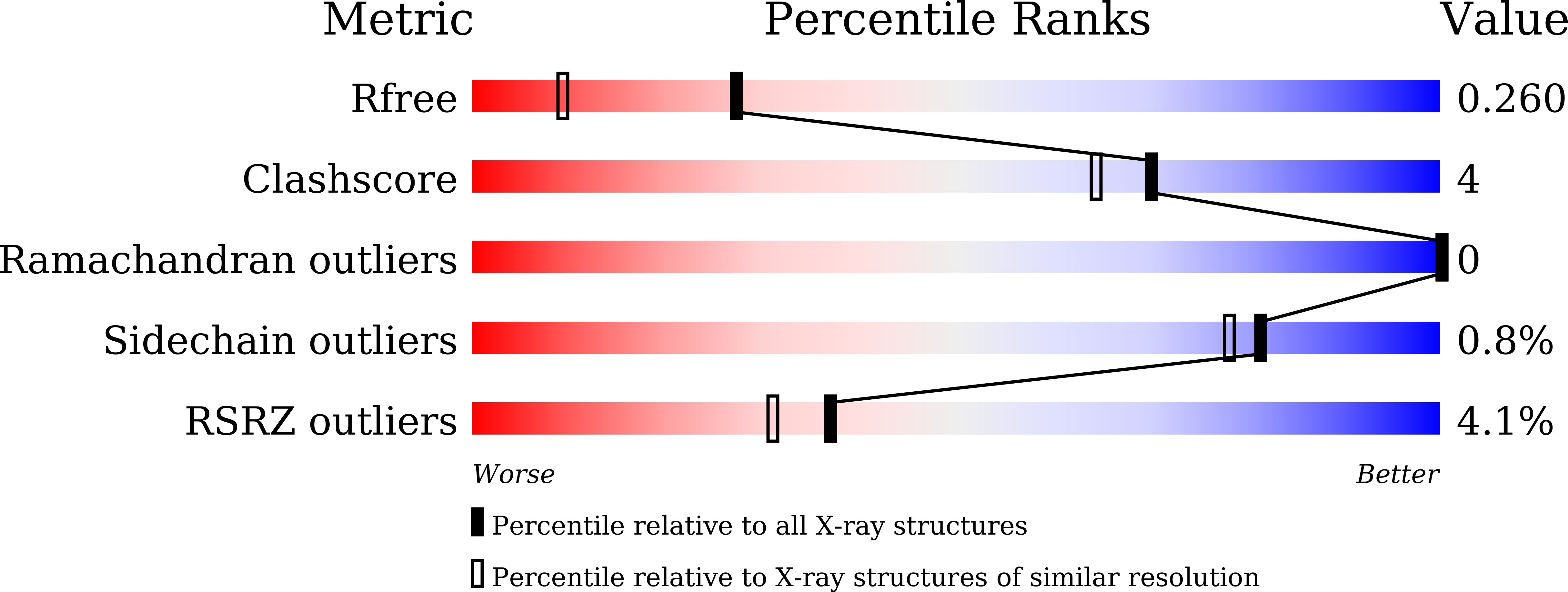
5X2L - PubMed Abstract:
Most of the endogenous free d-serine (about 90%) in the brain is produced by serine racemase (SR). d-Serine in the brain is involved in neurodegenerative disorders and epileptic states as an endogenous co-agonist of the NMDA-type glutamate receptor. Thus, SR inhibitors are expected to be novel therapeutic candidates for the treatment of these disorders. In this study, we solved the crystal structure of wild-type SR, and tried to identify a new inhibitor of SR by in silico screening using the structural information. As a result, we identified two hit compounds by their in vitro evaluations using wild-type SR. Based on the structure of the more potent hit compound 1, we synthesized 15 derivatives and evaluated their inhibitory activities against wild-type SR. Among them, the compound 9C showed relatively high inhibitory potency for wild-type SR. Compound 9C was a more potent inhibitor than compound 24, which was synthesized by our group based upon the structural information of the mutant-type SR.
Organizational Affiliation:
Graduate School of Innovative Life Science, University of Toyama, Japan.