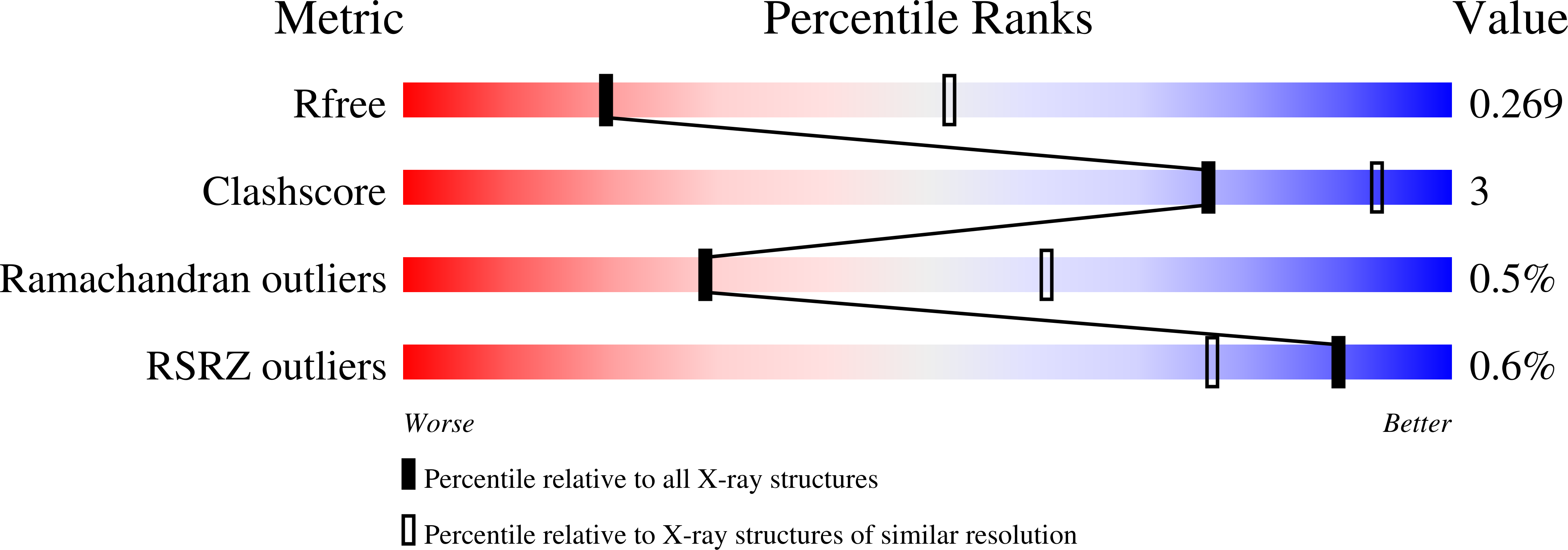
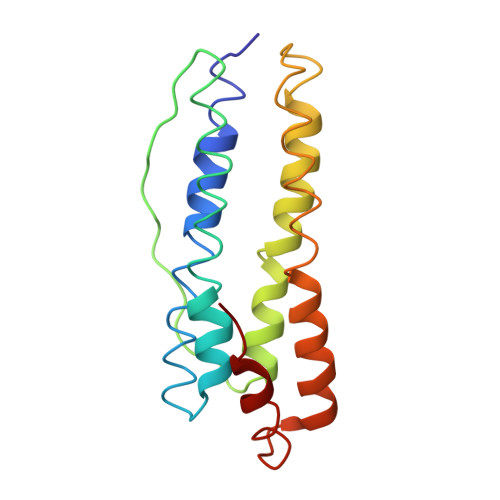
Thermophilic Ferritin 24mer Assembly and Nanoparticle Encapsulation Modulated by Interdimer Electrostatic Repulsion.
Pulsipher, K.W., Villegas, J.A., Roose, B.W., Hicks, T.L., Yoon, J., Saven, J.G., Dmochowski, I.J.(2017) Biochemistry 56: 3596-3606
- PubMed: 28682599
- DOI: https://doi.org/10.1021/acs.biochem.7b00296
- Primary Citation of Related Structures:
5V5K - PubMed Abstract:
Protein cage self-assembly enables encapsulation and sequestration of small molecules, macromolecules, and nanomaterials for many applications in bionanotechnology. Notably, wild-type thermophilic ferritin from Archaeoglobus fulgidus (AfFtn) exists as a stable dimer of four-helix bundle proteins at a low ionic strength, and the protein forms a hollow assembly of 24 protomers at a high ionic strength (∼800 mM NaCl). This assembly process can also be initiated by highly charged gold nanoparticles (AuNPs) in solution, leading to encapsulation. These data suggest that salt solutions or charged AuNPs can shield unfavorable electrostatic interactions at AfFtn dimer-dimer interfaces, but specific "hot-spot" residues controlling assembly have not been identified. To investigate this further, we computationally designed three AfFtn mutants (E65R, D138K, and A127R) that introduce a single positive charge at sites along the dimer-dimer interface. These proteins exhibited different assembly kinetics and thermodynamics, which were ranked in order of increasing 24mer propensity: A127R < wild type < D138K ≪ E65R. E65R assembled into the 24mer across a wide range of ionic strengths (0-800 mM NaCl), and the dissociation temperature for the 24mer was 98 °C. X-ray crystal structure analysis of the E65R mutant identified a more compact, closed-pore cage geometry. A127R and D138K mutants exhibited wild-type ability to encapsulate and stabilize 5 nm AuNPs, whereas E65R did not encapsulate AuNPs at the same high yields. This work illustrates designed protein cages with distinct assembly and encapsulation properties.
Organizational Affiliation:
Department of Chemistry, University of Pennsylvania , 231 South 34th Street, Philadelphia, Pennsylvania 19104, United States.