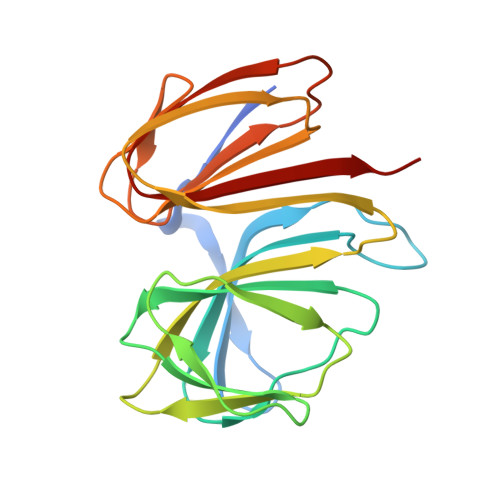
Crystal structure of a bicupin protein HutD involved in histidine utilization in Pseudomonas.
Gerth, M.L., Liu, Y., Jiao, W., Zhang, X.X., Baker, E.N., Lott, J.S., Rainey, P.B., Johnston, J.M.(2017) Proteins 85: 1580-1588
- PubMed: 28383128
- DOI: https://doi.org/10.1002/prot.25303
- Primary Citation of Related Structures:
5V00 - PubMed Abstract:
Cupins form one of the most functionally diverse superfamilies of proteins, with members performing a wide range of catalytic, non-catalytic, and regulatory functions. HutD is a predicted bicupin protein that is involved in histidine utilization (Hut) in Pseudomonas species. Previous genetic analyses have suggested that it limits the upper level of Hut pathway expression, but its mechanism of action is unknown. Here, we have determined the structure of PfluHutD at 1.74 Å resolution in several crystallization conditions, and identified N-formyl-l-glutamate (FG, a Hut pathway intermediate) as a potential ligand in vivo. Proteins 2017; 85:1580-1588. © 2017 Wiley Periodicals, Inc.
Organizational Affiliation:
New Zealand Institute for Advanced Study, Massey University Albany, Auckland, New Zealand.