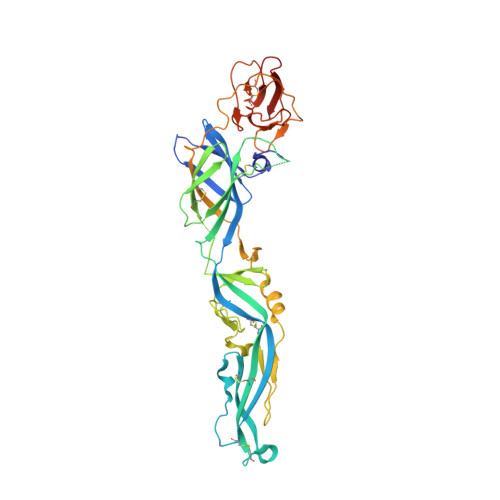
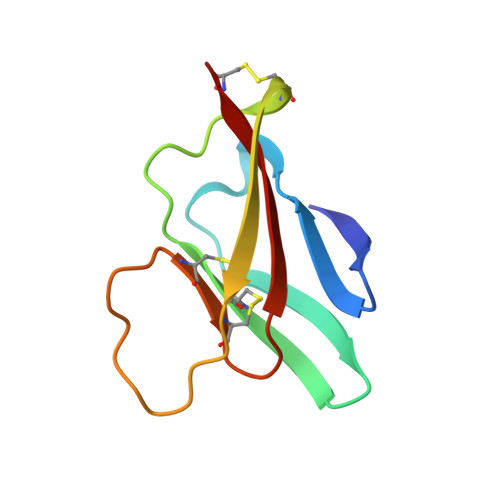
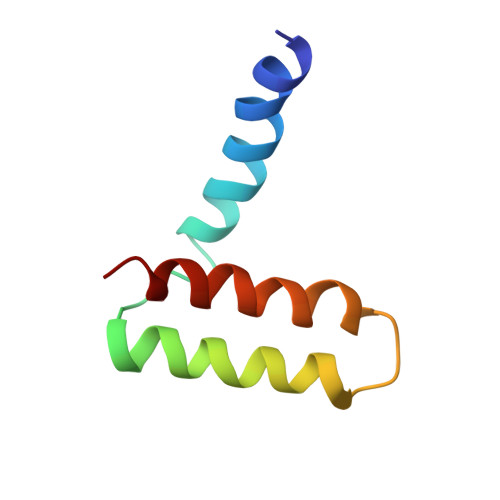
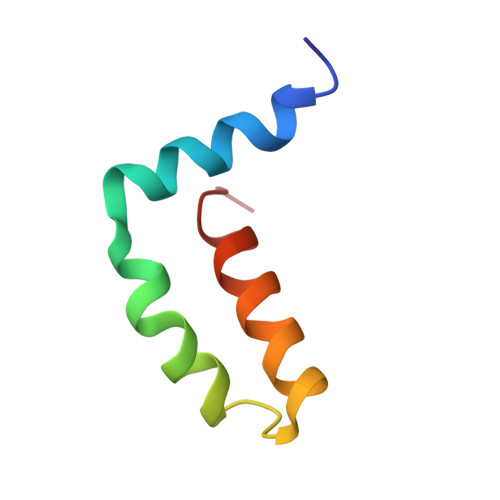
Structure of the immature Zika virus at 9 angstrom resolution.
Prasad, V.M., Miller, A.S., Klose, T., Sirohi, D., Buda, G., Jiang, W., Kuhn, R.J., Rossmann, M.G.(2017) Nat Struct Mol Biol 24: 184-186
- PubMed: 28067914
- DOI: https://doi.org/10.1038/nsmb.3352
- Primary Citation of Related Structures:
5U4W - PubMed Abstract:
The current Zika virus (ZIKV) epidemic is characterized by severe pathogenicity in both children and adults. Sequence changes in ZIKV since its first isolation are apparent when pre-epidemic strains are compared with those causing the current epidemic. However, the residues that are responsible for ZIKV pathogenicity are largely unknown. Here we report the cryo-electron microscopy (cryo-EM) structure of the immature ZIKV at 9-Å resolution. The cryo-EM map was fitted with the crystal structures of the precursor membrane and envelope glycoproteins and was shown to be similar to the structures of other known immature flaviviruses. However, the immature ZIKV contains a partially ordered capsid protein shell that is less prominent in other immature flaviviruses. Furthermore, six amino acids near the interface between pr domains at the top of the spikes were found to be different between the pre-epidemic and epidemic ZIKV, possibly influencing the composition and structure of the resulting viruses.
Organizational Affiliation:
Department of Biological Sciences, Purdue University, West Lafayette, Indiana, USA.