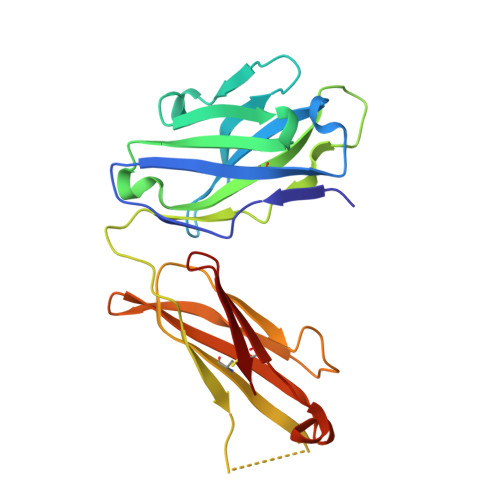
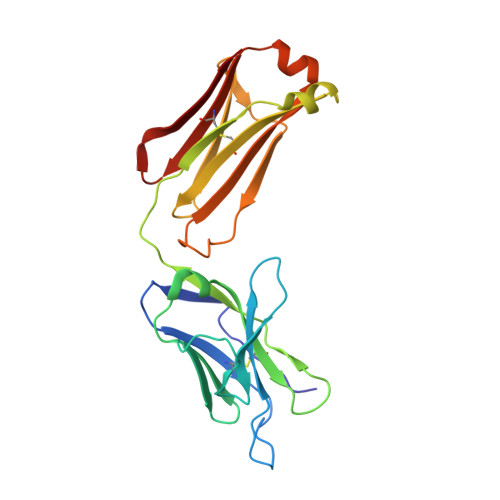
Neisseria meningitidis factor H-binding protein bound to monoclonal antibody JAR5: implications for antibody synergy.
Malito, E., Lo Surdo, P., Veggi, D., Santini, L., Stefek, H., Brunelli, B., Luzzi, E., Bottomley, M.J., Beernink, P.T., Scarselli, M.(2016) Biochem J 473: 4699-4713
- PubMed: 27784765
- DOI: https://doi.org/10.1042/BCJ20160806
- Primary Citation of Related Structures:
5T5F - PubMed Abstract:
Factor H-binding protein (fHbp) is an important antigen of Neisseria meningitidis that is capable of eliciting a robust protective immune response in humans. Previous studies on the interactions of fHbp with antibodies revealed that some anti-fHbp monoclonal antibodies that are unable to trigger complement-mediated bacterial killing in vitro are highly co-operative and become bactericidal if used in combination. Several factors have been shown to influence such co-operativity, including IgG subclass and antigen density. To investigate the structural basis of the anti-fHbp antibody synergy, we determined the crystal structure of the complex between fHbp and the Fab (fragment antigen-binding) fragment of JAR5, a specific anti-fHbp murine monoclonal antibody known to be highly co-operative with other monoclonal antibodies. We show that JAR5 is highly synergic with monoclonal antibody (mAb) 12C1, whose structure in complex with fHbp has been previously solved. Structural analyses of the epitopes recognized by JAR5 and 12C1, and computational modeling of full-length IgG mAbs of JAR5 and 12C1 bound to the same fHbp molecule, provide insights into the spatial orientation of Fc (fragment crystallizable) regions and into the possible implications for the susceptibility of meningococci to complement-mediated killing.
Organizational Affiliation:
GSK Vaccines srl, Via Fiorentina 1, Siena 53100, Italy.