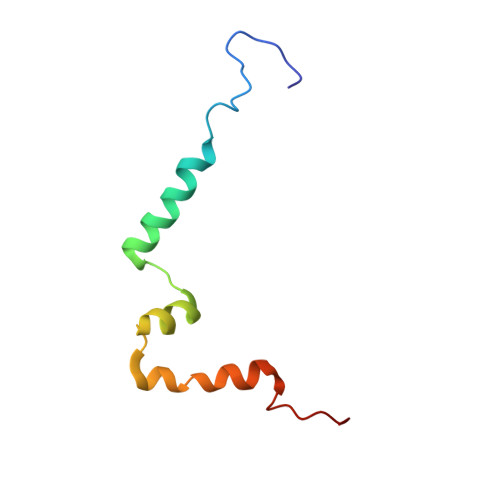
Structure of the competence pilus major pilin ComGC in Streptococcus pneumoniae.
Muschiol, S., Erlendsson, S., Aschtgen, M.S., Oliveira, V., Schmieder, P., de Lichtenberg, C., Teilum, K., Boesen, T., Akbey, U., Henriques-Normark, B.(2017) J Biol Chem 292: 14134-14146
- PubMed: 28659339
- DOI: https://doi.org/10.1074/jbc.M117.787671
- Primary Citation of Related Structures:
5NCA - PubMed Abstract:
Type IV pili are important virulence factors on the surface of many pathogenic bacteria and have been implicated in a wide range of diverse functions, including attachment, twitching motility, biofilm formation, and horizontal gene transfer. The respiratory pathogen Streptococcus pneumoniae deploys type IV pili to take up DNA during transformation. These "competence pili" are composed of the major pilin protein ComGC and exclusively assembled during bacterial competence, but their biogenesis remains unclear. Here, we report the high resolution NMR structure of N-terminal truncated ComGC revealing a highly flexible and structurally divergent type IV pilin. It consists of only three α-helical segments forming a well-defined electronegative cavity and confined electronegative and hydrophobic patches. The structure is particularly flexible between the first and second α-helix with the first helical part exhibiting slightly slower dynamics than the rest of the pilin, suggesting that the first helix is involved in forming the pilus structure core and that parts of helices two and three are primarily surface-exposed. Taken together, our results provide the first structure of a type IV pilin protein involved in the formation of competence-induced pili in Gram-positive bacteria and corroborate the remarkable structural diversity among type IV pilin proteins.
Organizational Affiliation:
From the Department of Microbiology, Tumor and Cell Biology, Karolinska Institutet, 171 77 Stockholm, Sweden,; Department of Clinical Microbiology, Karolinska University Hospital, 171 76 Stockholm, Sweden,. Electronic address: sandra.muschiol@ki.se.