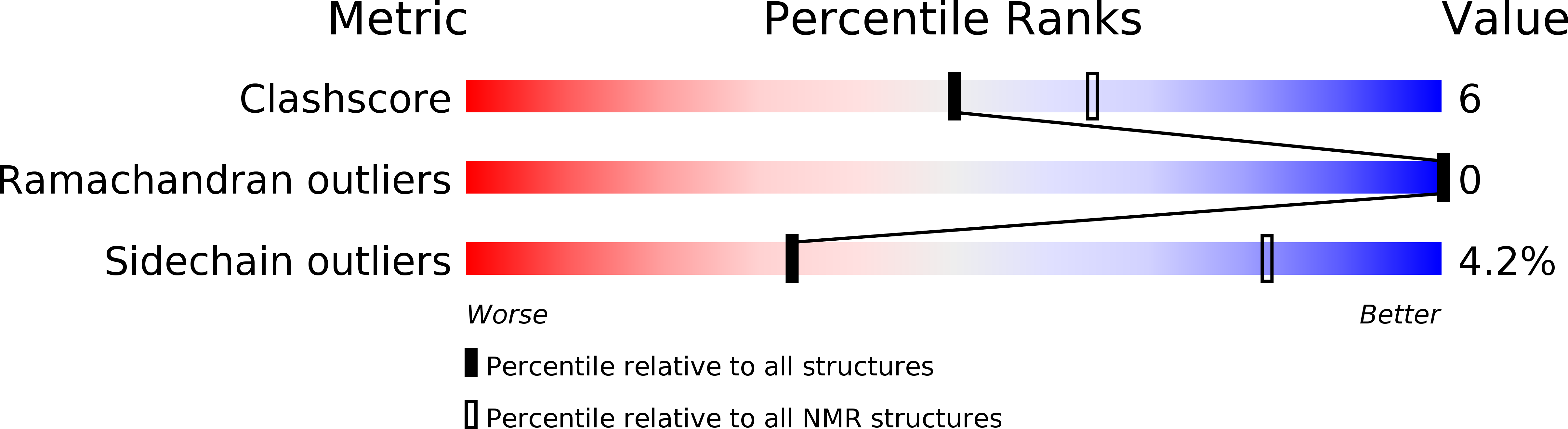Distinct roles of Pcf11 zinc-binding domains in pre-mRNA 3'-end processing.
Guegueniat, J., Dupin, A.F., Stojko, J., Beaurepaire, L., Cianferani, S., Mackereth, C.D., Minvielle-Sebastia, L., Fribourg, S.(2017) Nucleic Acids Res 45: 10115-10131
- PubMed: 28973460
- DOI: https://doi.org/10.1093/nar/gkx674
- Primary Citation of Related Structures:
5M9Z - PubMed Abstract:
New transcripts generated by RNA polymerase II (RNAPII) are generally processed in order to form mature mRNAs. Two key processing steps include a precise cleavage within the 3' end of the pre-mRNA, and the subsequent polymerization of adenosines to produce the poly(A) tail. In yeast, these two functions are performed by a large multi-subunit complex that includes the Cleavage Factor IA (CF IA). The four proteins Pcf11, Clp1, Rna14 and Rna15 constitute the yeast CF IA, and of these, Pcf11 is structurally the least characterized. Here, we provide evidence for the binding of two Zn2+ atoms to Pcf11, bound to separate zinc-binding domains located on each side of the Clp1 recognition region. Additional structural characterization of the second zinc-binding domain shows that it forms an unusual zinc finger fold. We further demonstrate that the two domains are not mandatory for CF IA assembly nor RNA polymerase II transcription termination, but are rather involved to different extents in the pre-mRNA 3'-end processing mechanism. Our data thus contribute to a more complete understanding of the architecture and function of Pcf11 and its role within the yeast CF IA complex.
Organizational Affiliation:
Université de Bordeaux, INSERM U1212, CNRS UMR5320, Bordeaux, France.