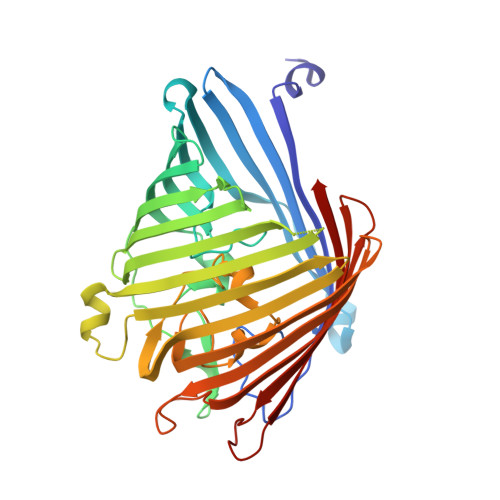
MOMP from Campylobacter jejuni Is a Trimer of 18-Stranded beta-Barrel Monomers with a Ca(2+) Ion Bound at the Constriction Zone.
Ferrara, L.G., Wallat, G.D., Moynie, L., Dhanasekar, N.N., Aliouane, S., Acosta-Gutierrez, S., Pages, J.M., Bolla, J.M., Winterhalter, M., Ceccarelli, M., Naismith, J.H.(2016) J Mol Biol 428: 4528-4543
- PubMed: 27693650
- DOI: https://doi.org/10.1016/j.jmb.2016.09.021
- Primary Citation of Related Structures:
5LDT, 5LDV - PubMed Abstract:
The Gram-negative organism Campylobacter jejuni is the major cause of food poisoning. Unlike Escherichia coli, which has two major porins, OmpC and OmpF, C. jejuni has one, termed major outer membrane protein (MOMP) through which nutrients and antibiotics transit. We report the 2.1-Å crystal structure of C. jejuni MOMP expressed in E. coli and a lower resolution but otherwise identical structure purified directly from C. jejuni. The 2.1-Å resolution structure of recombinant MOMP showed that although the protein has timeric arrangement similar to OmpC, it is an 18-stranded, not 16-stranded, β-barrel. The structure has identified a Ca 2+ bound at the constriction zone, which is functionally significant as suggested by molecular dynamics and single-channel experiments. The water-filled channel of MOMP has a narrow constriction zone, and single-molecule studies show a monomeric conductivity of 0.7±0.2 nS and a trimeric conductance of 2.2±0.2 nS. The ion neutralizes negative charges at the constriction zone, reducing the transverse electric field and reversing ion selectivity. Modeling of the transit of ciprofloxacin, an antibiotic of choice for treating Campylobacter infection, through the pore of MOMP reveals a trajectory that is dependent upon the presence metal ion.
Organizational Affiliation:
Biomedical Sciences Research Complex, University of St Andrews, 09042 St Andrews, UK.