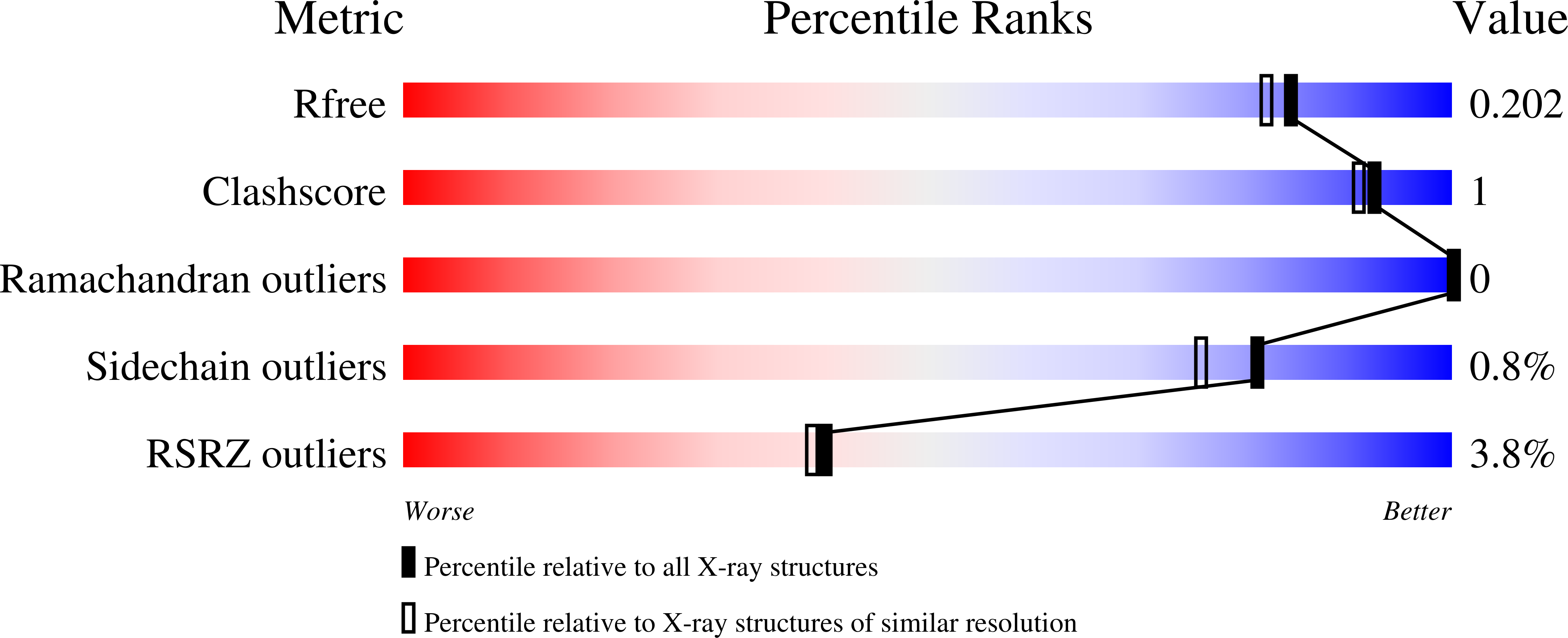
The structure of a doripenem-bound OXA-51 class D beta-lactamase variant with enhanced carbapenemase activity.
June, C.M., Muckenthaler, T.J., Schroder, E.C., Klamer, Z.L., Wawrzak, Z., Powers, R.A., Szarecka, A., Leonard, D.A.(2016) Protein Sci 25: 2152-2163
- PubMed: 27636561
- DOI: https://doi.org/10.1002/pro.3040
- Primary Citation of Related Structures:
5KZH, 5L2F - PubMed Abstract:
OXA-51 is a class D β-lactamase that is thought to be the native carbapenemase of Acinetobacter baumannii. Many variants of OXA-51 containing active site substitutions have been identified from A. baumannii isolates, and some of these substitutions increase hydrolytic activity toward carbapenem antibiotics. We have determined the high-resolution structures of apo OXA-51 and OXA-51 with one such substitution (I129L) with the carbapenem doripenem trapped in the active site as an acyl-intermediate. The structure shows that acyl-doripenem adopts an orientation very similar to carbapenem ligands observed in the active site of OXA-24/40 (doripenem) and OXA-23 (meropenem). In the OXA-51 variant/doripenem complex, the indole ring of W222 is oriented away from the doripenem binding site, thereby eliminating a clash that is predicted to occur in wildtype OXA-51. Similarly, in the OXA-51 variant complex, L129 adopts a different rotamer compared to I129 in wildtype OXA-51. This alternative position moves its side chain away from the hydroxyethyl moiety of doripenem and relieves another potential clash between the enzyme and carbapenem substrates. Molecular dynamics simulations of OXA-51 and OXA-51 I129L demonstrate that compared to isoleucine, a leucine at this position greatly favors a rotamer that accommodates the ligand. These results provide a molecular justification for how this substitution generates enhanced binding affinity for carbapenems, and therefore helps explain the prevalence of this substitution in clinical OXA-51 variants.
Organizational Affiliation:
Department of Chemistry, Grand Valley State University, Allendale, Michigan, 49401.