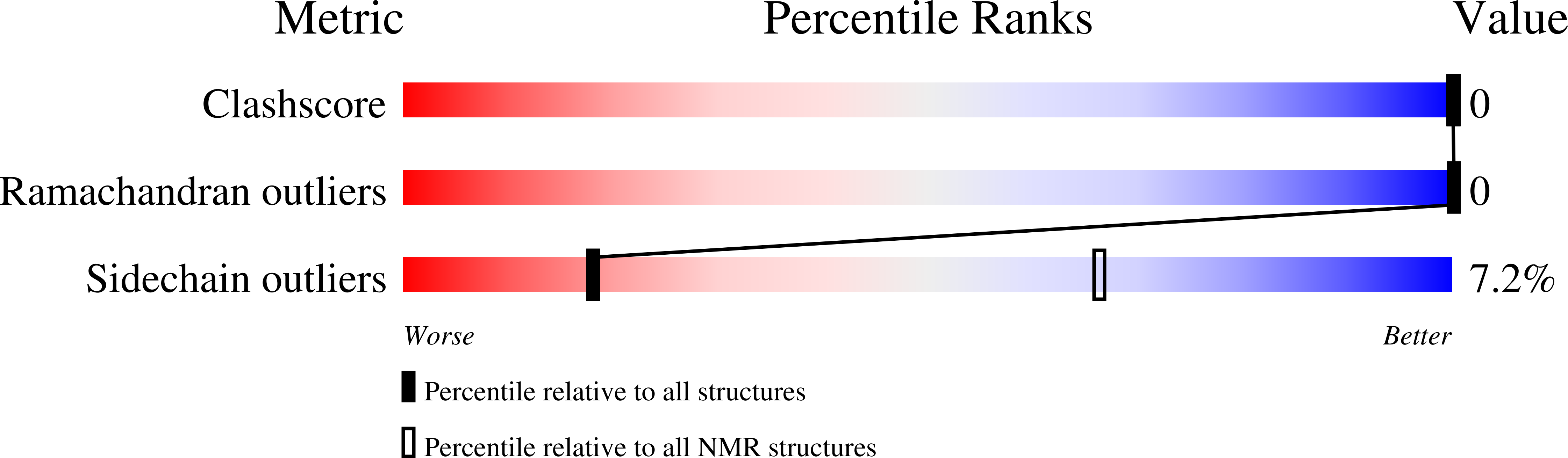Identification and structural characterization of LytU, a unique peptidoglycan endopeptidase from the lysostaphin family.
Raulinaitis, V., Tossavainen, H., Aitio, O., Juuti, J.T., Hiramatsu, K., Kontinen, V., Permi, P.(2017) Sci Rep 7: 6020-6020
- PubMed: 28729697
- DOI: https://doi.org/10.1038/s41598-017-06135-w
- Primary Citation of Related Structures:
5KQB, 5KQC - PubMed Abstract:
We introduce LytU, a short member of the lysostaphin family of zinc-dependent pentaglycine endopeptidases. It is a potential antimicrobial agent for S. aureus infections and its gene transcription is highly upregulated upon antibiotic treatments along with other genes involved in cell wall synthesis. We found this enzyme to be responsible for the opening of the cell wall peptidoglycan layer during cell divisions in S. aureus. LytU is anchored in the plasma membrane with the active part residing in the periplasmic space. It has a unique Ile/Lys insertion at position 151 that resides in the catalytic site-neighbouring loop and is vital for the enzymatic activity but not affecting the overall structure common to the lysostaphin family. Purified LytU lyses S. aureus cells and cleaves pentaglycine, a reaction conveniently monitored by NMR spectroscopy. Substituting the cofactor zinc ion with a copper or cobalt ion remarkably increases the rate of pentaglycine cleavage. NMR and isothermal titration calorimetry further reveal that, uniquely for its family, LytU is able to bind a second zinc ion which is coordinated by catalytic histidines and is therefore inhibitory. The pH-dependence and high affinity of binding carry further physiological implications.
Organizational Affiliation:
Program in Structural Biology and Biophysics, Institute of Biotechnology, University of Helsinki, Viikinkaari 1, P.O. Box 65, FI-00014, Helsinki, Finland.