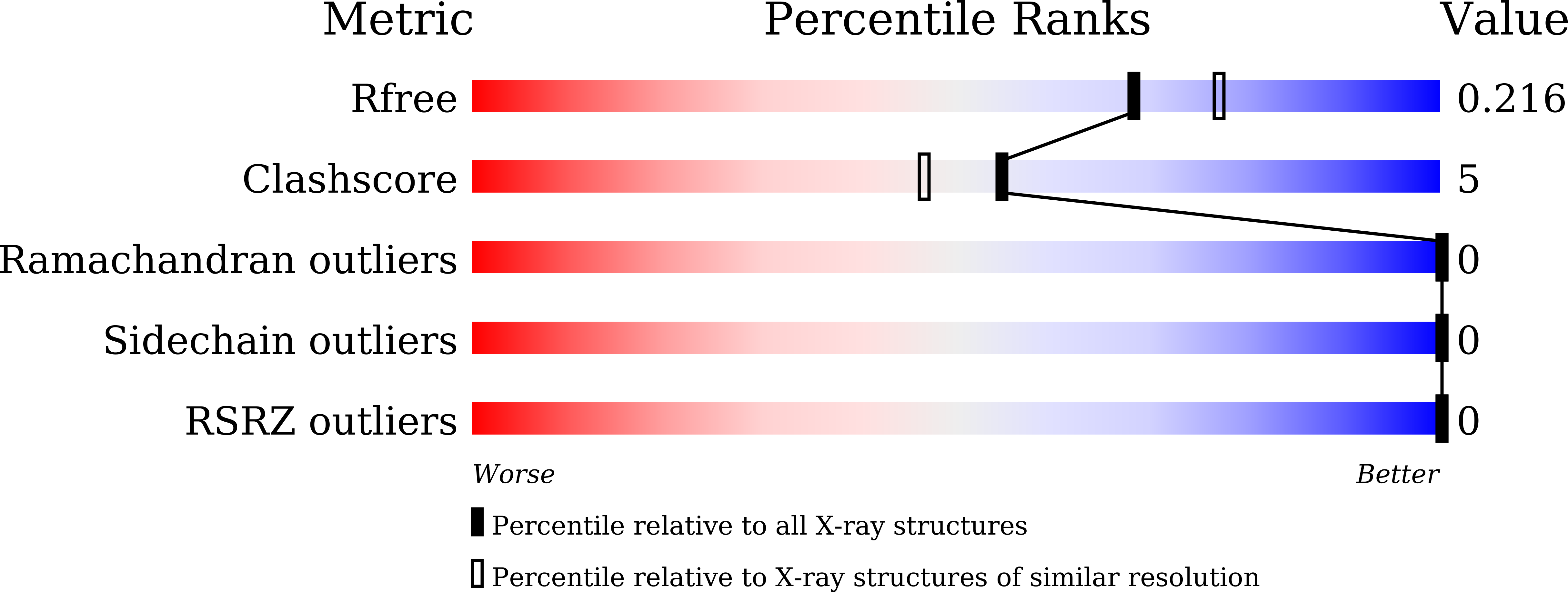
Structural insights into the binding of buckwheat glutaredoxin with GSH and regulation of its catalytic activity
Zhang, X., Wang, W., Li, C., Zhao, Y., Yuan, H., Tan, X., Wu, L., Wang, Z., Wang, H.(2017) J Inorg Biochem 173: 21-27
- PubMed: 28478310
- DOI: https://doi.org/10.1016/j.jinorgbio.2017.04.019
- Primary Citation of Related Structures:
5GTX, 5KQA - PubMed Abstract:
Glutaredoxins (Grxs) are ubiquitous thioltransferases and members of the thioredoxin (Trx) fold superfamily. They have multiple functions in cells including oxidative stress responses and cell signaling. A novel glutaredoxin from buckwheat (rbGrx) with higher catalytic activity was identified, cloned, and purified. The structures of glutathionylated rbGrx and an rbGrx mutant, in which cysteine 39 was mutated to alanine, were solved by x-ray diffraction at a resolution of 2.05Å and 2.29Å, respectively. In rbGrx, GSH (glutathione) is bound at the conserved GSH-binding site, and its structure shows that it has the potential to function as a scaffold protein for the assembly and delivery of GSH. The crystal structure shows that GSH does not bind to the C39A rbGrx mutant, and the C39A mutant had no catalytic activity, indicating that C39 is a key residue that is involved in both the binding of rbGrx to GSH and the regulation of its catalytic activity. The model showing the binding of GSH with rbGrx provides a basis for understanding its molecular function and its potential future applications in medicinal food science.
Organizational Affiliation:
College of Life Science, Shanxi University, Shanxi, Taiyuan 030006, China.