Interrupting peptidoglycan deacetylation during Bdellovibrio predator-prey interaction prevents ultimate destruction of prey wall, liberating bacterial-ghosts.
Lambert, C., Lerner, T.R., Bui, N.K., Somers, H., Aizawa, S., Liddell, S., Clark, A., Vollmer, W., Lovering, A.L., Sockett, R.E.(2016) Sci Rep 6: 26010-26010
- PubMed: 27211869
- DOI: https://doi.org/10.1038/srep26010
- Primary Citation of Related Structures:
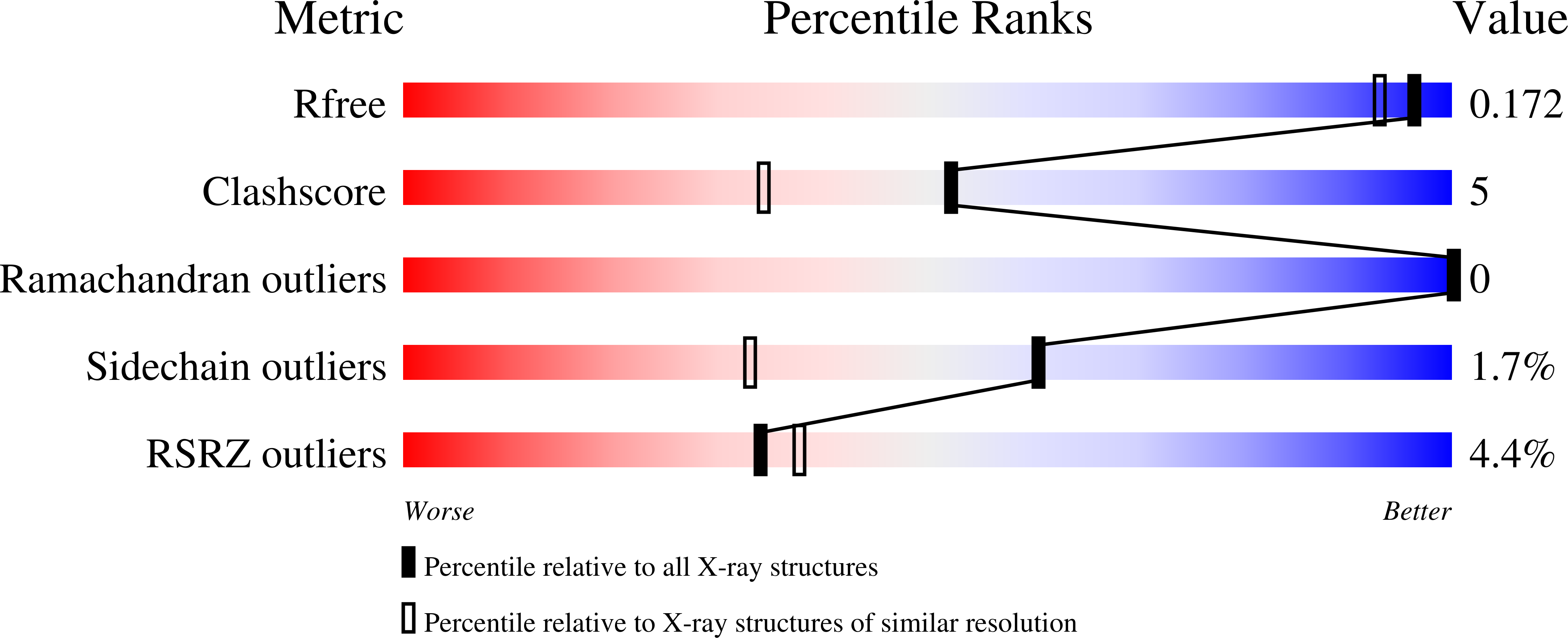
5JP6 - PubMed Abstract:
The peptidoglycan wall, located in the periplasm between the inner and outer membranes of the cell envelope in Gram-negative bacteria, maintains cell shape and endows osmotic robustness. Predatory Bdellovibrio bacteria invade the periplasm of other bacterial prey cells, usually crossing the peptidoglycan layer, forming transient structures called bdelloplasts within which the predators replicate. Prey peptidoglycan remains intact for several hours, but is modified and then degraded by escaping predators. Here we show predation is altered by deleting two Bdellovibrio N-acetylglucosamine (GlcNAc) deacetylases, one of which we show to have a unique two domain structure with a novel regulatory"plug". Deleting the deacetylases limits peptidoglycan degradation and rounded prey cell "ghosts" persist after mutant-predator exit. Mutant predators can replicate unusually in the periplasmic region between the peptidoglycan wall and the outer membrane rather than between wall and inner-membrane, yet still obtain nutrients from the prey cytoplasm. Deleting two further genes encoding DacB/PBP4 family proteins, known to decrosslink and round prey peptidoglycan, results in a quadruple mutant Bdellovibrio which leaves prey-shaped ghosts upon predation. The resultant bacterial ghosts contain cytoplasmic membrane within bacteria-shaped peptidoglycan surrounded by outer membrane material which could have promise as "bacterial skeletons" for housing artificial chromosomes.
Organizational Affiliation:
Centre for Genetics and Genomics, School of Life Sciences, University of Nottingham, Medical School, Queen's Medical Centre, Nottingham, NG7 2UH, UK.