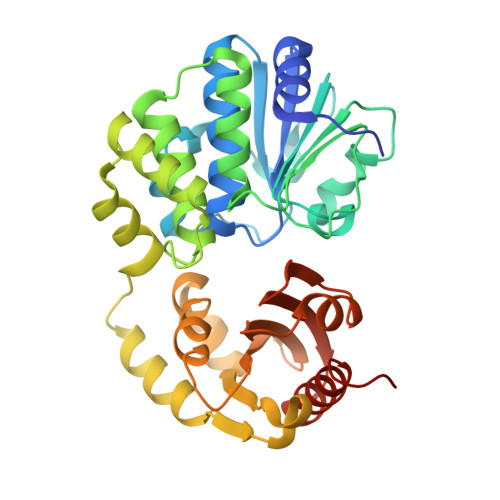
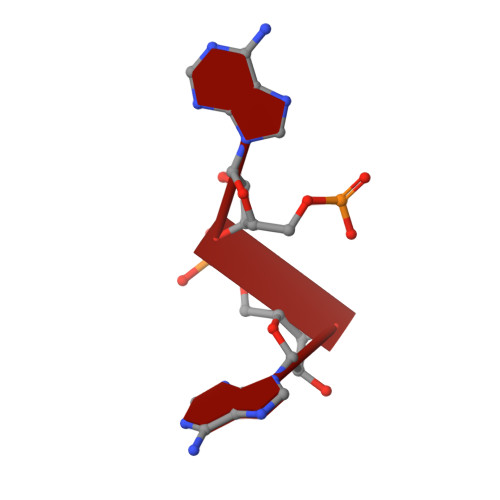
Structural and biochemical insight into the mechanism of Rv2837c from Mycobacterium tuberculosis as a c-di-NMP phosphodiesterase
He, Q., Wang, F., Liu, S., Zhu, D., Cong, H., Gao, F., Li, B., Wang, H., Lin, Z., Liao, J., Gu, L.(2016) J Biol Chem 291: 14386-14387
- PubMed: 27371563
- DOI: https://doi.org/10.1074/jbc.A115.699801
- Primary Citation of Related Structures:
5JJU