Structural analysis of beta-prism lectin from Colocasia esculenta (L.) S chott.
Vajravijayan, S., Pletnev, S., Pletnev, V.Z., Nandhagopal, N., Gunasekaran, K.(2016) Int J Biol Macromol 91: 518-523
- PubMed: 27262515
- DOI: https://doi.org/10.1016/j.ijbiomac.2016.05.048
- Primary Citation of Related Structures:
5J76 - PubMed Abstract:
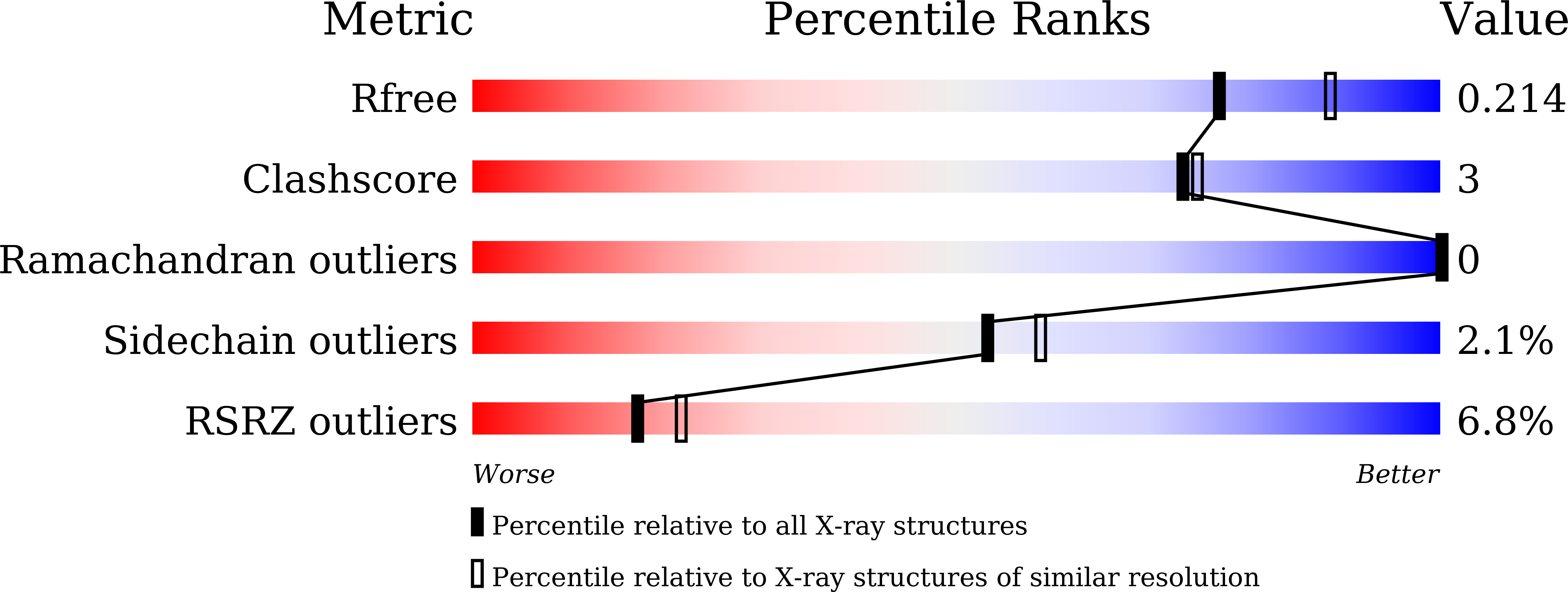
The Mannose-binding β-Prism Colocasia esculenta lectin (β-PCL) was purified from tubers using ion exchange chromatography. The purified β-PCL appeared as a single band of ∼12kDa on SDS-PAGE. β-PCL crystallizes in trigonal space group P3121 and diffracted to a resolution of 2.1Å. The structure was solved using Molecular replacement using Crocus vernus lectin (PDB: 3MEZ) as a model. From the final refined model to an R-factor of 16.5% and an Rfree of 20.4%, it has been observed that the biological unit consists of two β-Prism domains augmented through C-terminals swap over to form one of faces for each domain. Cα superposition of individual domains of β-PCL with individual domains of other related structures and superposition of whole protein structures were carried out. The higher RMS deviation for the superposition of whole structures suggest that β-prism domains assume different orientation in each structure.
Organizational Affiliation:
Centre of Advanced Study in Crystallography and Biophysics, University of Madras, Guindy Campus, Chennai 600 025, India.