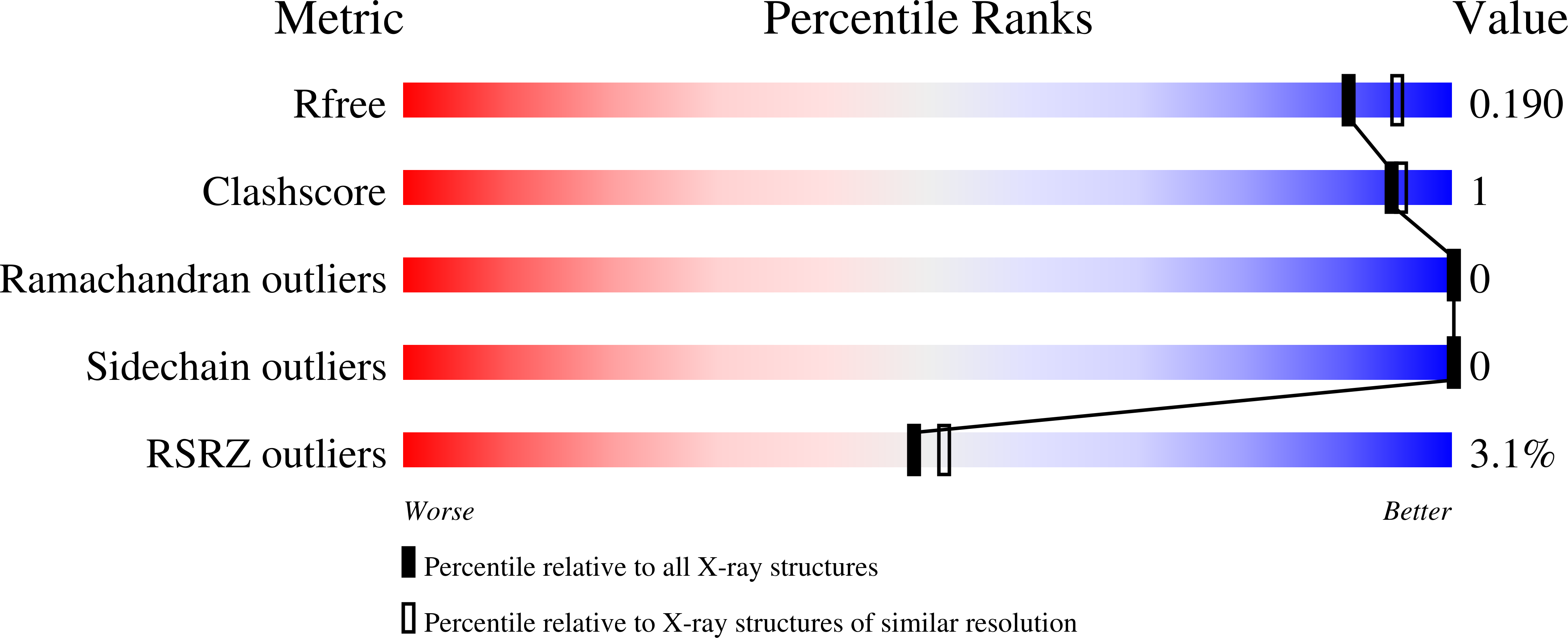
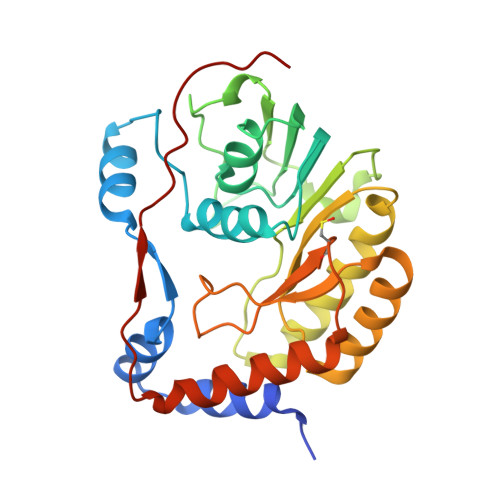
1.9 Angstrom Crystal Structure of NS5 Methyl Transferase from Dengue Virus 1 in Complex with S-Adenosylmethionine and Beta-D-Fructopyranose.
Minasov, G., Shuvalova, L., Winsor, J., Dubrovska, I., Flores, K., Shatsman, S., Kwon, K., Anderson, W.F., Center for Structural Genomics of Infectious DiseasesTo be published.