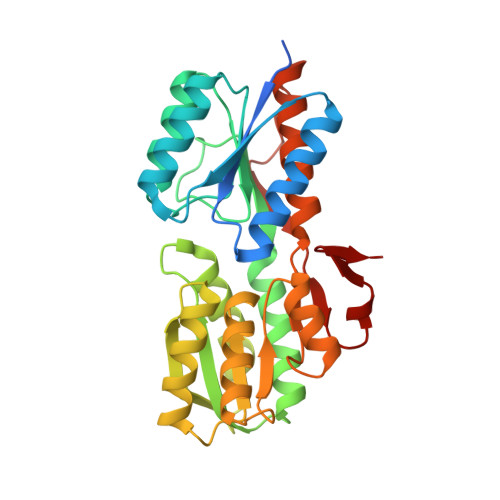
Crystal structure of an ABC solute binding protein from Rhizobium etli CFN 42 (RHE_PF00037,TARGET EFI-511357) in complex with alpha-D-apiose
Vetting, M.W., Carter, M.S., Al Obaidi, N.F., Morisco, L.L., Benach, J., Koss, J., Wasserman, S.R., Gerlt, J.A., Almo, S.C., Enzyme Function Initiative (EFI)To be published.