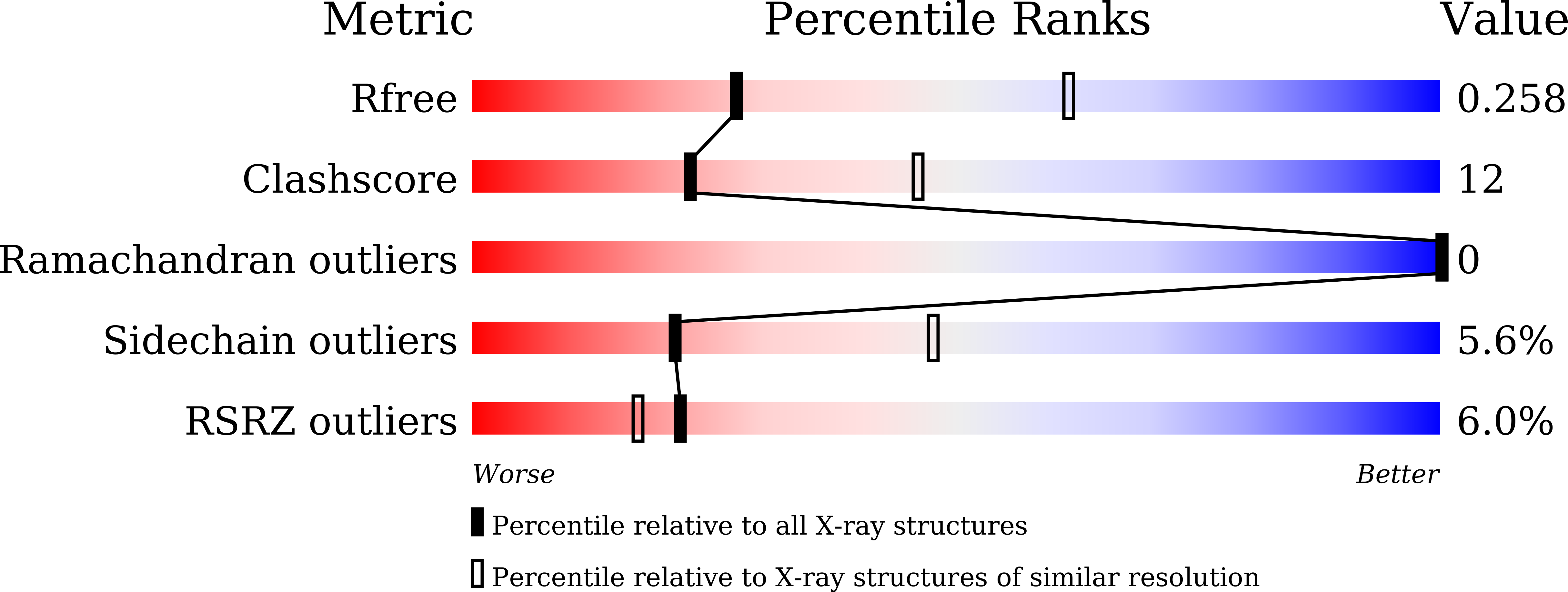
A Mutation Directs the Structural Switch of DNA Binding Proteins under Starvation to a Ferritin-like Protein Cage.
Williams, S.M., Chandran, A.V., Prakash, S., Vijayan, M., Chatterji, D.(2017) Structure 25: 1449-1454.e3
- PubMed: 28823472
- DOI: https://doi.org/10.1016/j.str.2017.07.006
- Primary Citation of Related Structures:
5H46 - PubMed Abstract:
Proteins of the ferritin family are ubiquitous in living organisms. With their spherical cage-like structures they are the iron storehouses in cells. Subfamilies of ferritins include 24-meric ferritins and bacterioferritins (maxiferritins), and 12-meric Dps (miniferritins). Dps safeguards DNA by direct binding, affording physical protection and safeguards from free radical-mediated damage by sequestering iron in its core. The maxiferritins can oxidize and store iron but cannot bind DNA. Here we show that a mutation at a critical interface in Dps alters its assembly from the canonical 12-mer to a ferritin-like 24-mer under crystallization. This structural switch was attributed to the conformational alteration of a highly conserved helical loop and rearrangement of the C-terminus. Our results demonstrate a novel concept of mutational switch between related protein subfamilies and corroborate the popular model for evolution by which subtle substitutions in an amino acid sequence lead to diversification among proteins.
Organizational Affiliation:
Molecular Biophysics Unit, Indian Institute of Science, Bangalore 560 012, India.