The Immunity-Related Gtpase Irga6 Dimerizes in a Parallel Head-to-Head Fashion.
Schulte, K., Pawlowski, N., Faelber, K., Froehlich, C., Howard, J., Daumke, O.(2016) BMC Biol 14: 14
- PubMed: 26934976
- DOI: https://doi.org/10.1186/s12915-016-0236-7
- Primary Citation of Related Structures:
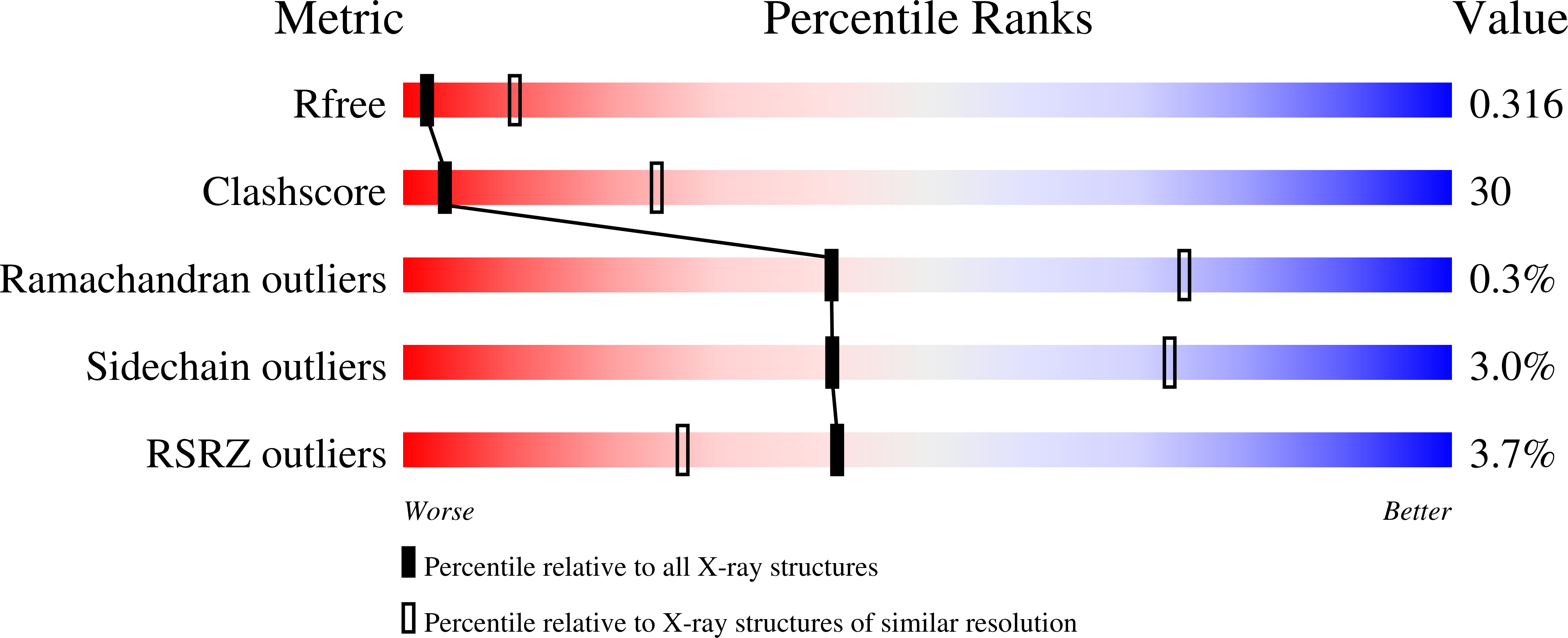
5FPH - PubMed Abstract:
The immunity-related GTPases (IRGs) constitute a powerful cell-autonomous resistance system against several intracellular pathogens. Irga6 is a dynamin-like protein that oligomerizes at the parasitophorous vacuolar membrane (PVM) of Toxoplasma gondii leading to its vesiculation. Based on a previous biochemical analysis, it has been proposed that the GTPase domains of Irga6 dimerize in an antiparallel fashion during oligomerization.
Organizational Affiliation:
Max-Delbrueck-Centrum for Molecular Medicine, Crystallography, Robert-Rössle-Strasse 10, 13125, Berlin, Germany.