Discovery of a proteinaceous cellular receptor for a norovirus.
Orchard, R.C., Wilen, C.B., Doench, J.G., Baldridge, M.T., McCune, B.T., Lee, Y.C., Lee, S., Pruett-Miller, S.M., Nelson, C.A., Fremont, D.H., Virgin, H.W.(2016) Science 353: 933-936
- PubMed: 27540007
- DOI: https://doi.org/10.1126/science.aaf1220
- Primary Citation of Related Structures:
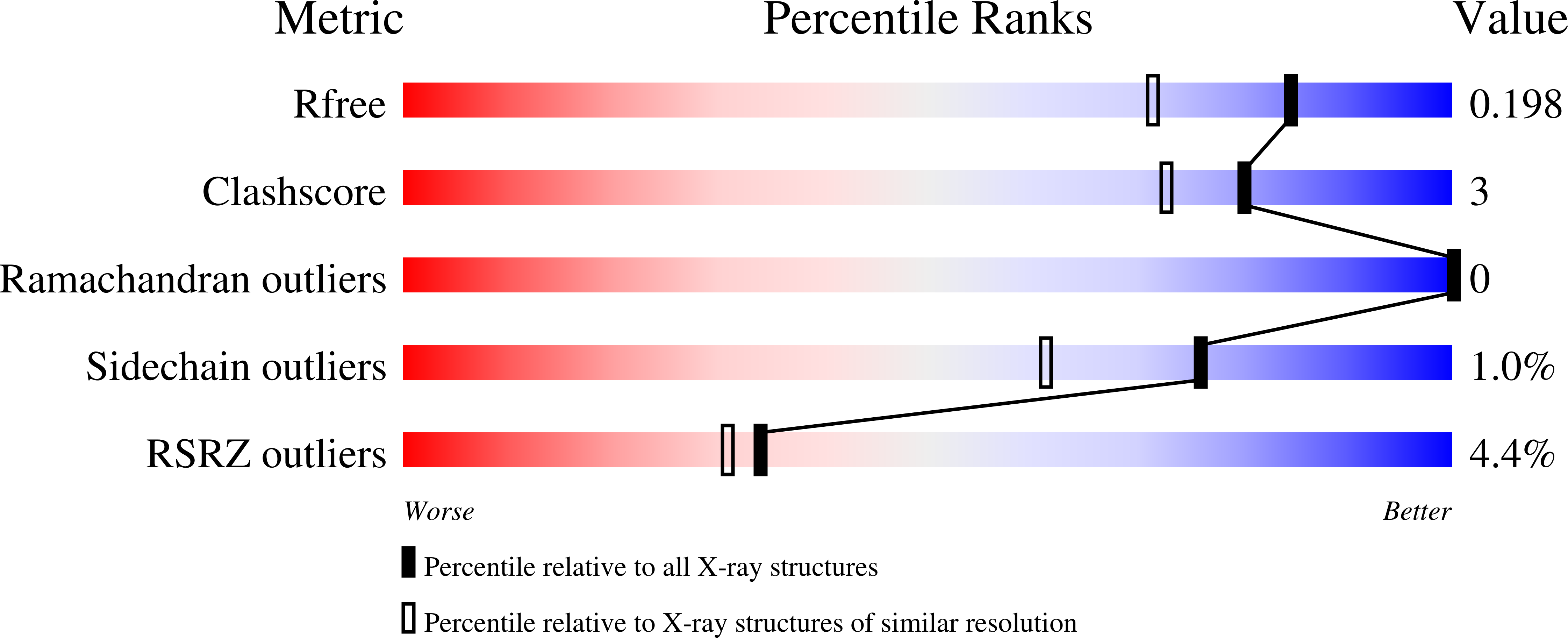
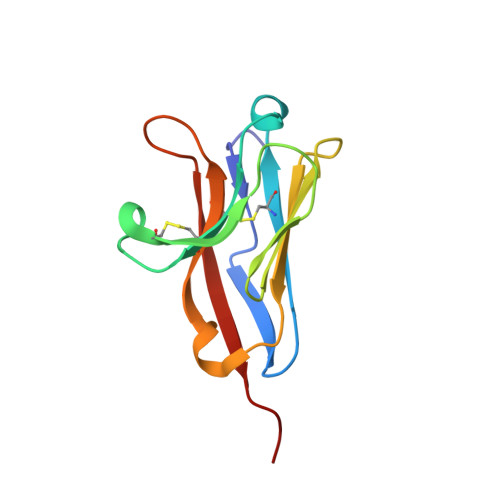
5FFL - PubMed Abstract:
Noroviruses (NoVs) are a leading cause of gastroenteritis globally, yet the host factors required for NoV infection are poorly understood. We identified host molecules that are essential for murine NoV (MNoV)-induced cell death, including CD300lf as a proteinaceous receptor. We found that CD300lf is essential for MNoV binding and replication in cell lines and primary cells. Additionally, Cd300lf(-/-) mice are resistant to MNoV infection. Expression of CD300lf in human cells breaks the species barrier that would otherwise restrict MNoV replication. The crystal structure of the CD300lf ectodomain reveals a potential ligand-binding cleft composed of residues that are critical for MNoV infection. Therefore, the presence of a proteinaceous receptor is the primary determinant of MNoV species tropism, whereas other components of cellular machinery required for NoV replication are conserved between humans and mice.
Organizational Affiliation:
Department of Pathology and Immunology, Washington University School of Medicine, St. Louis, MO 63110, USA.