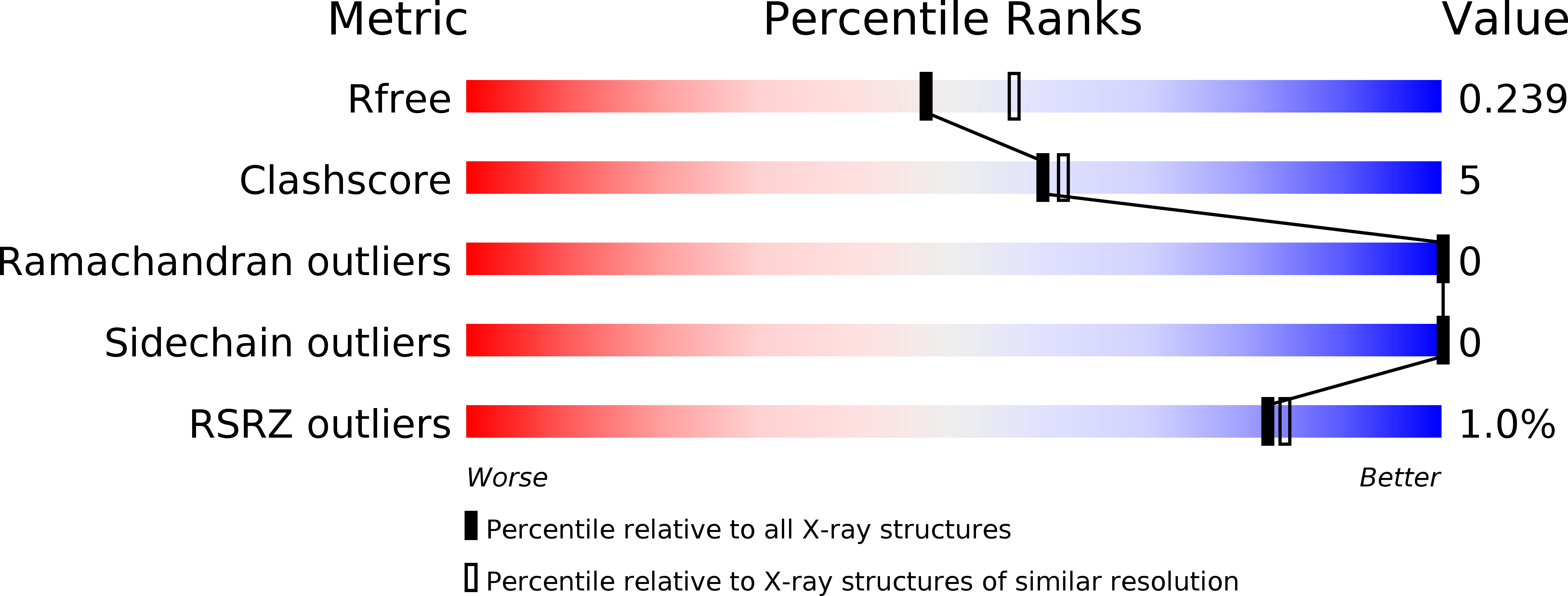
Structural analysis of Centrolobium tomentosum seed lectin with inflammatory activity.
Almeida, A.C., Osterne, V.J., Santiago, M.Q., Pinto-Junior, V.R., Silva-Filho, J.C., Lossio, C.F., Nascimento, F.L., Almeida, R.P., Teixeira, C.S., Leal, R.B., Delatorre, P., Rocha, B.A., Assreuy, A.M., Nascimento, K.S., Cavada, B.S.(2016) Arch Biochem Biophys 596: 73-83
- PubMed: 26946944
- DOI: https://doi.org/10.1016/j.abb.2016.03.001
- Primary Citation of Related Structures:
5EYX, 5EYY - PubMed Abstract:
A glycosylated lectin (CTL) with specificity for mannose and glucose has been detected and purified from seeds of Centrolobium tomentosum, a legume plant from Dalbergieae tribe. It was isolated by mannose-sepharose affinity chromatography. The primary structure was determined by tandem mass spectrometry and consists of 245 amino acids, similar to other Dalbergieae lectins. CTL structures were solved from two crystal forms, a monoclinic and a tetragonal, diffracted at 2.25 and 1.9 Å, respectively. The carbohydrate recognition domain (CRD), metal-binding site and glycosylation site were characterized, and the structural basis for mannose/glucose-binding was elucidated. The lectin adopts the canonical dimeric organization of legume lectins. CTL showed acute inflammatory effect in paw edema model. The protein was subjected to ligand screening (dimannosides and trimannoside) by molecular docking, and interactions were compared with similar lectins possessing the same ligand specificity. This is the first crystal structure of mannose/glucose native seed lectin with proinflammatory activity isolated from the Centrolobium genus.
Organizational Affiliation:
Laboratório de Moléculas Biologicamente Ativas - BioMol-Lab, Departamento de Bioquímica e Biologia Molecular, Universidade Federal do Ceará, Fortaleza, Ceará, Brazil.