Molecular and Structural Characterization of a Novel Escherichia coli Interleukin Receptor Mimic Protein.
Moriel, D.G., Heras, B., Paxman, J.J., Lo, A.W., Tan, L., Sullivan, M.J., Dando, S.J., Beatson, S.A., Ulett, G.C., Schembri, M.A.(2016) mBio 7: e02046-e02046
- PubMed: 26980835
- DOI: https://doi.org/10.1128/mBio.02046-15
- Primary Citation of Related Structures:
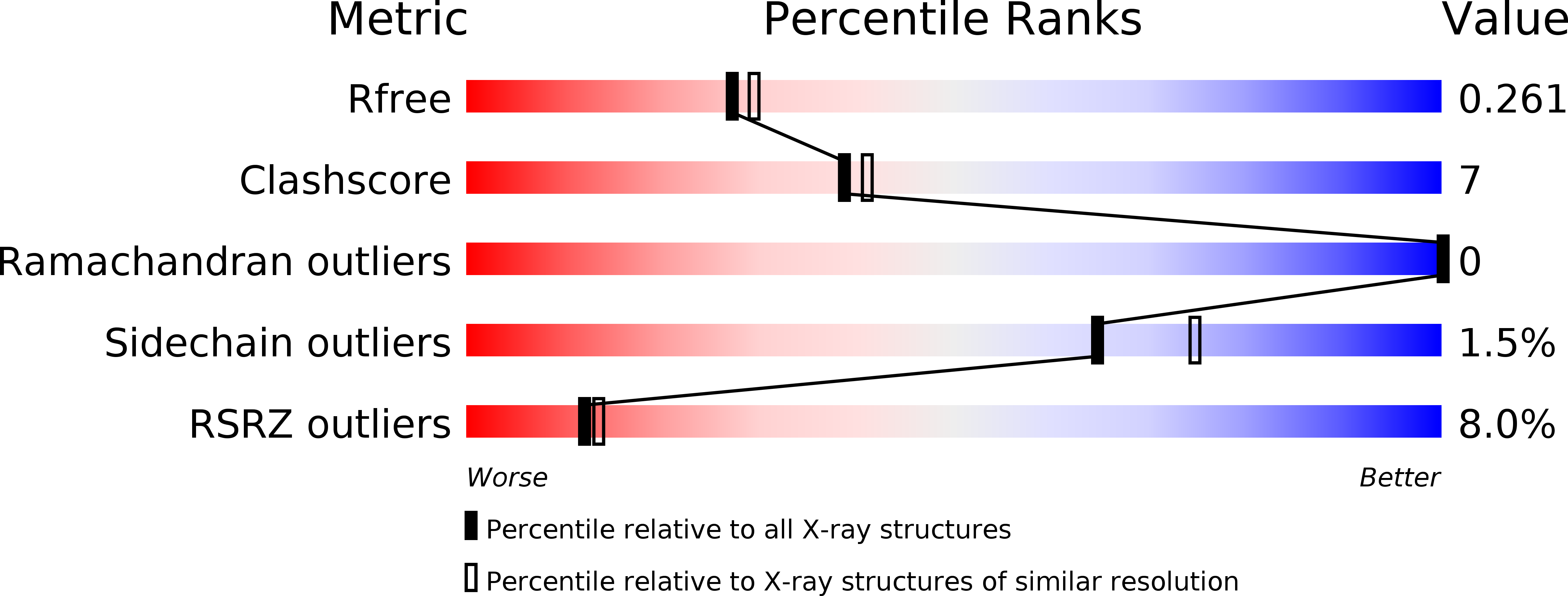
5EK5 - PubMed Abstract:
Urinary tract infection (UTI) is a disease of extremely high incidence in both community and nosocomial settings. UTIs cause significant morbidity and mortality, with approximately 150 million cases globally per year. Uropathogenic Escherichia coli (UPEC) is the primary cause of UTI and is generally treated empirically. However, the rapidly increasing incidence of UTIs caused by multidrug-resistant UPEC strains has led to limited available treatment options and highlights the urgent need to develop alternative treatment and prevention strategies. In this study, we performed a comprehensive analysis to define the regulation, structure, function, and immunogenicity of recently identified UPEC vaccine candidate C1275 (here referred to as IrmA). We showed that the irmA gene is highly prevalent in UPEC, is cotranscribed with the biofilm-associated antigen 43 gene, and is regulated by the global oxidative stress response OxyR protein. Localization studies identified IrmA in the UPEC culture supernatant. We determined the structure of IrmA and showed that it adopts a unique domain-swapped dimer architecture. The dimeric structure of IrmA displays similarity to those of human cytokine receptors, including the interleukin-2 receptor (IL-2R), interleukin-4 receptor (IL-4R), and interleukin-10 receptor (IL-10R) binding domains, and we showed that purified IrmA can bind to their cognate cytokines. Finally, we showed that plasma from convalescent urosepsis patients contains high IrmA antibody titers, demonstrating the strong immunogenicity of IrmA. Taken together, our results indicate that IrmA may play an important role during UPEC infection.
Organizational Affiliation:
Australian Infectious Diseases Research Centre, School of Chemistry and Molecular Biosciences, The University of Queensland, Brisbane, Australia.