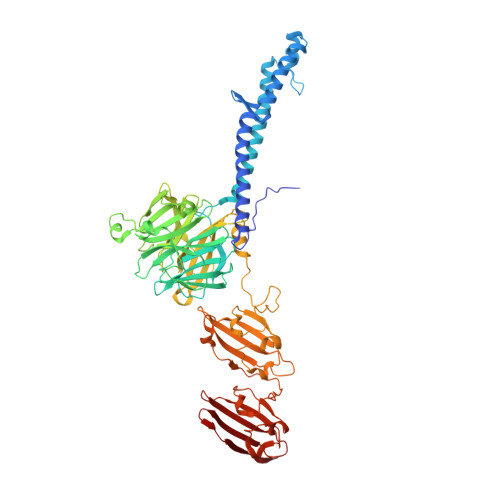
Structure of the host-recognition device of Staphylococcus aureus phage 11.
Koc, C., Xia, G., Kuhner, P., Spinelli, S., Roussel, A., Cambillau, C., Stehle, T.(2016) Sci Rep 6: 27581-27581
- PubMed: 27282779
- DOI: https://doi.org/10.1038/srep27581
- Primary Citation of Related Structures:
5EFV - PubMed Abstract:
Phages play key roles in the pathogenicity and adaptation of the human pathogen Staphylococcus aureus. However, little is known about the molecular recognition events that mediate phage adsorption to the surface of S. aureus. The lysogenic siphophage ϕ11 infects S. aureus SA113. It was shown previously that ϕ11 requires α- or β-N-acetylglucosamine (GlcNAc) moieties on cell wall teichoic acid (WTA) for adsorption. Gp45 was identified as the receptor binding protein (RBP) involved in this process and GlcNAc residues on WTA were found to be the key component of the ϕ11 receptor. Here we report the crystal structure of the RBP of ϕ11, which assembles into a large, multidomain homotrimer. Each monomer contains a five-bladed propeller domain with a cavity that could accommodate a GlcNAc moiety. An electron microscopy reconstruction of the ϕ11 host adhesion component, the baseplate, reveals that six RBP trimers are assembled around the baseplate core. The Gp45 and baseplate structures provide insights into the overall organization and molecular recognition process of the phage ϕ11 tail. This assembly is conserved among most glycan-recognizing Siphoviridae, and the RBP orientation would allow host adhesion and infection without an activation step.
Organizational Affiliation:
Interfaculty Institute of Biochemistry, University of Tübingen, 72076 Tübingen, Germany.