Secondary PDZ domain-binding site on class B plexins enhances the affinity for PDZ-RhoGEF.
Pascoe, H.G., Gutowski, S., Chen, H., Brautigam, C.A., Chen, Z., Sternweis, P.C., Zhang, X.(2015) Proc Natl Acad Sci U S A 112: 14852-14857
- PubMed: 26627240
- DOI: https://doi.org/10.1073/pnas.1508931112
- Primary Citation of Related Structures:
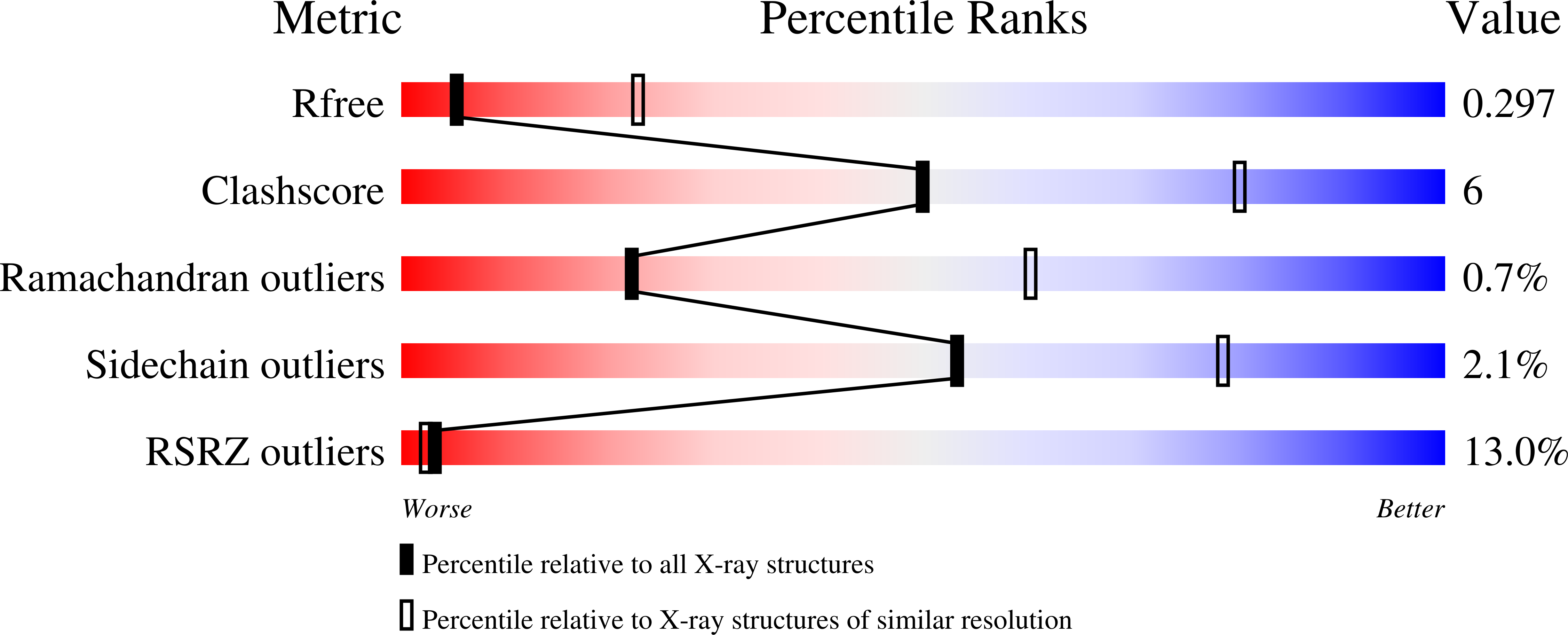
5E6P - PubMed Abstract:
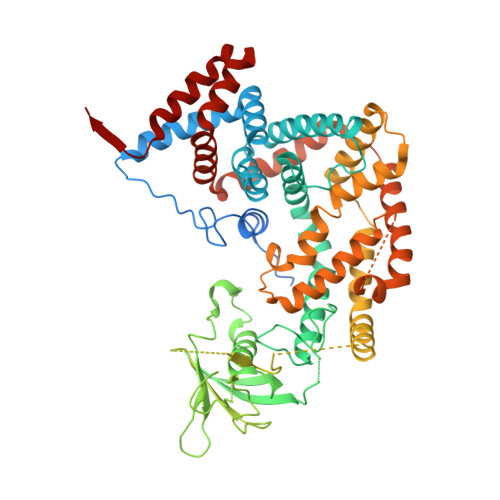
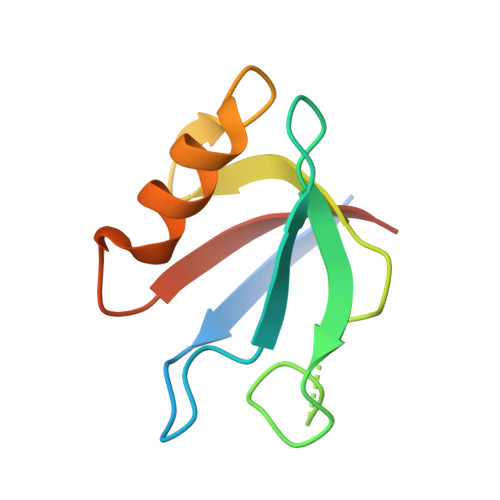
PDZ domains are abundant protein interaction modules and typically recognize a short motif at the C terminus of their ligands, with a few residues in the motif endowing the binding specificity. The sequence-based rules, however, cannot fully account for the specificity between the vast number of PDZ domains and ligands in the cell. Plexins are transmembrane receptors that regulate processes such as axon guidance and angiogenesis. Two related guanine nucleotide exchange factors (GEFs), PDZ-RhoGEF and leukemia-associated RhoGEF (LARG), use their PDZ domains to bind class B plexins and play critical roles in signaling. Here, we present the crystal structure of the full-length cytoplasmic region of PlexinB2 in complex with the PDZ domain of PDZ-RhoGEF. The structure reveals that, in addition to the canonical C-terminal motif/PDZ interaction, the 3D domain of PlexinB2 forms a secondary interface with the PDZ domain. Our biophysical and cell-based assays show that the secondary interface contributes to the specific interaction between plexin and PDZ-RhoGEF and to signaling by plexin in the cell. Formation of secondary interfaces may be a general mechanism for increasing affinity and specificity of modular domain-mediated interactions.
Organizational Affiliation:
Department of Pharmacology, University of Texas Southwestern Medical Center, Dallas, TX 75390;