Development and Characterization of Potent Cyclic Acyldepsipeptide Analogues with Increased Antimicrobial Activity.
Goodreid, J.D., Janetzko, J., Santa Maria, J.P., Wong, K.S., Leung, E., Eger, B.T., Bryson, S., Pai, E.F., Gray-Owen, S.D., Walker, S., Houry, W.A., Batey, R.A.(2016) J Med Chem 59: 624-646
- PubMed: 26818454
- DOI: https://doi.org/10.1021/acs.jmedchem.5b01451
- Primary Citation of Related Structures:
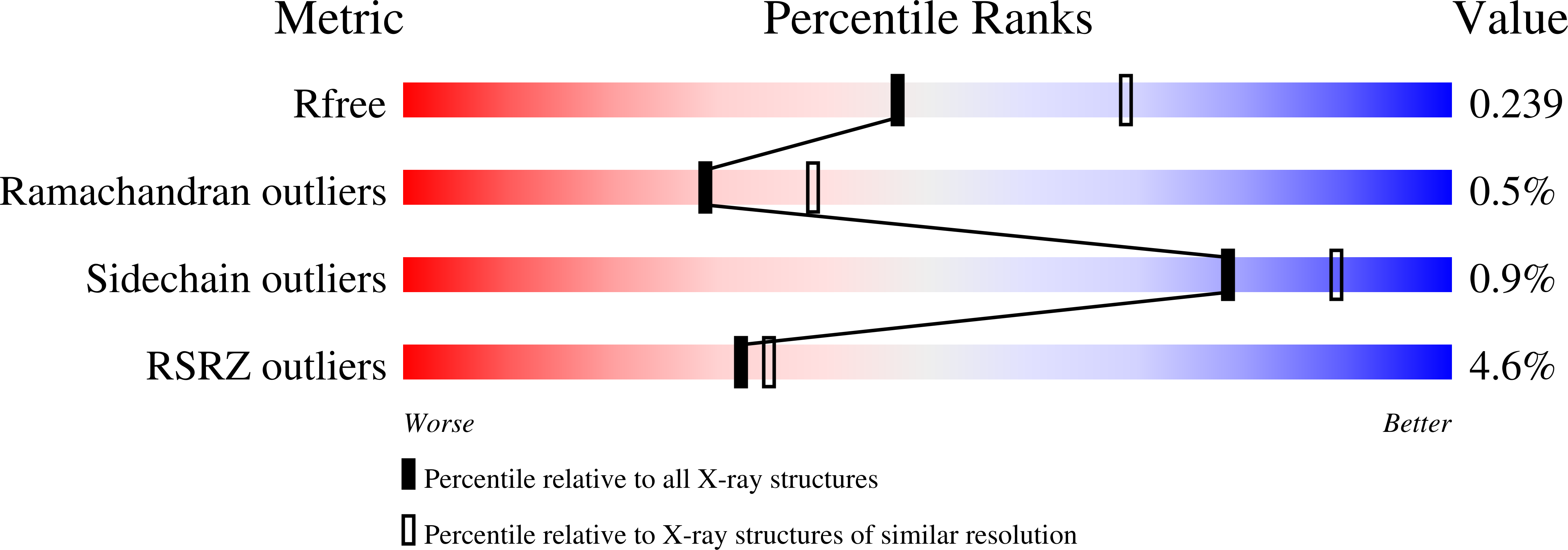
5DKP - PubMed Abstract:
The problem of antibiotic resistance has prompted the search for new antibiotics with novel mechanisms of action. Analogues of the A54556 cyclic acyldepsipeptides (ADEPs) represent an attractive class of antimicrobial agents that act through dysregulation of caseinolytic protease (ClpP). Previous studies have shown that ADEPs are active against Gram-positive bacteria (e.g., MRSA, VRE, PRSP (penicillin-resistant Streptococcus pneumoniae)); however, there are currently few studies examining Gram-negative bacteria. In this study, the synthesis and biological evaluation of 14 novel ADEPs against a variety of pathogenic Gram-negative and Gram-positive organisms is outlined. Optimization of the macrocyclic core residues and N-acyl side chain culminated in the development of 26, which shows potent activity against the Gram-negative species Neisseria meningitidis and Neisseria gonorrheae and improved activity against the Gram-positive organisms Staphylococcus aureus and Enterococcus faecalis in comparison with known analogues. In addition, the co-crystal structure of an ADEP-ClpP complex derived from N. meningitidis was solved.
Organizational Affiliation:
Davenport Research Laboratories, Department of Chemistry, University of Toronto , 80 St. George Street, Toronto, Ontario M5S 3H6, Canada.