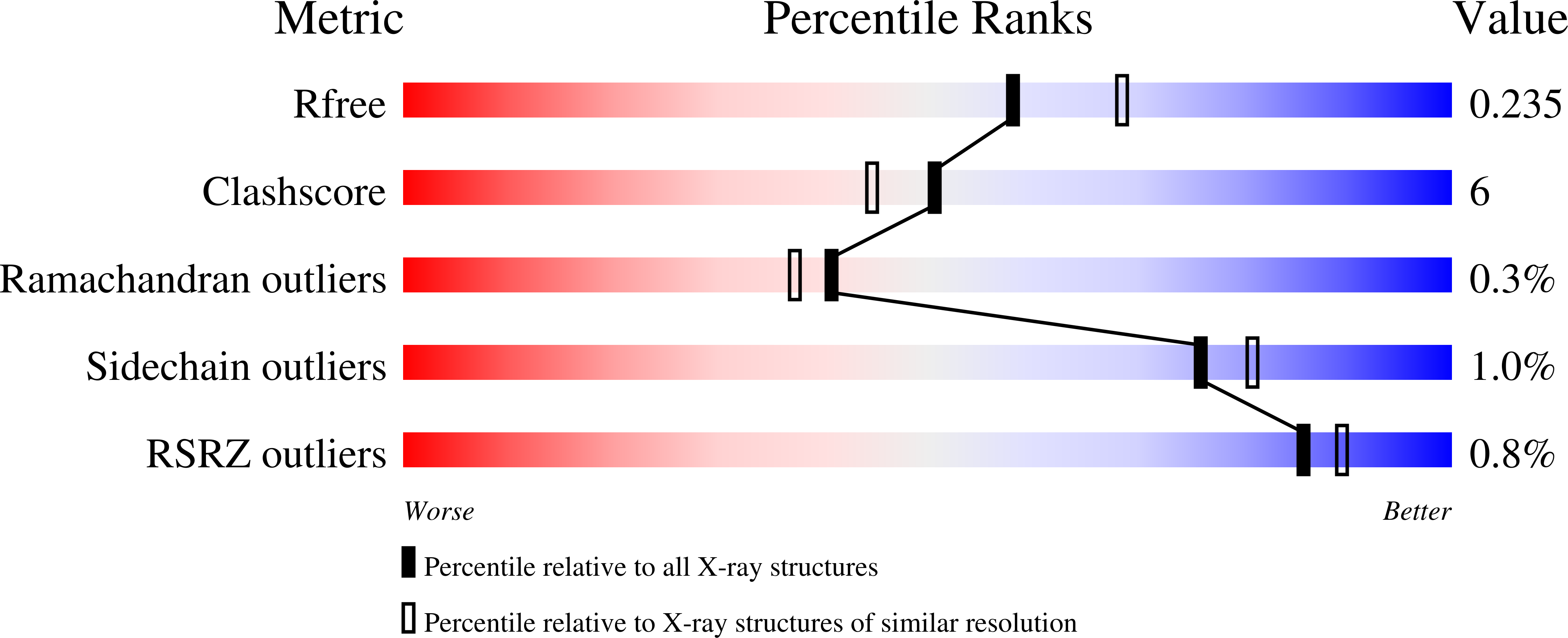
A Structural, Functional, and Computational Analysis of BshA, the First Enzyme in the Bacillithiol Biosynthesis Pathway.
Winchell, K.R., Egeler, P.W., VanDuinen, A.J., Jackson, L.B., Karpen, M.E., Cook, P.D.(2016) Biochemistry 55: 4654-4665
- PubMed: 27454321
- DOI: https://doi.org/10.1021/acs.biochem.6b00472
- Primary Citation of Related Structures:
5D00, 5D01 - PubMed Abstract:
Bacillithiol is a compound produced by several Gram-positive bacterial species, including the human pathogens Staphylococcus aureus and Bacillus anthracis. It is involved in maintaining cellular redox balance as well as the destruction of reactive oxygen species and harmful xenobiotic agents, including the antibiotic fosfomycin. BshA, BshB, and BshC are the enzymes involved in bacillithiol biosynthesis. BshA is a retaining glycosyltransferase responsible for the first committed step in bacillithiol production, namely the addition of N-acetylglucosamine to l-malate. Retaining glycosyltransferases like BshA are proposed to utilize an SNi-like reaction mechanism in which leaving group departure and nucleophilic attack occur on the same face of the hexose. However, significant questions regarding the details of how BshA and similar enzymes accommodate their substrates and facilitate catalysis persist. Here we report X-ray crystallographic structures of BshA from Bacillus subtilis 168 bound with UMP and/or GlcNAc-mal at resolutions of 2.15 and 2.02 Å, respectively. These ligand-bound structures, along with our functional and computational studies, provide clearer insight into how BshA and other retaining GT-B glycosyltransferases operate, corroborating the substrate-assisted, SNi-like reaction mechanism. The analyses presented herein can serve as the basis for the design of inhibitors capable of preventing bacillithiol production and, subsequently, help combat resistance to fosfomycin in various pathogenic Gram-positive microorganisms.
Organizational Affiliation:
Department of Chemistry, Grand Valley State University , Allendale, Michigan 49401, United States.