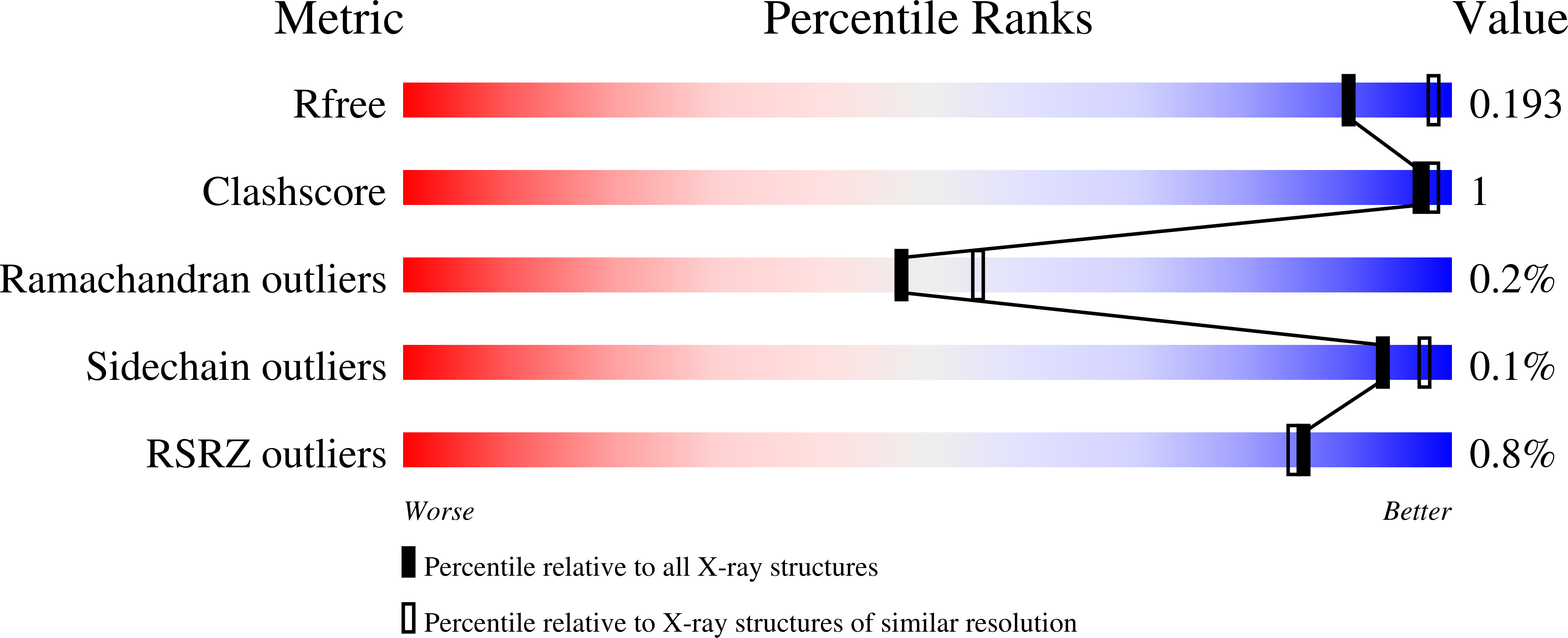
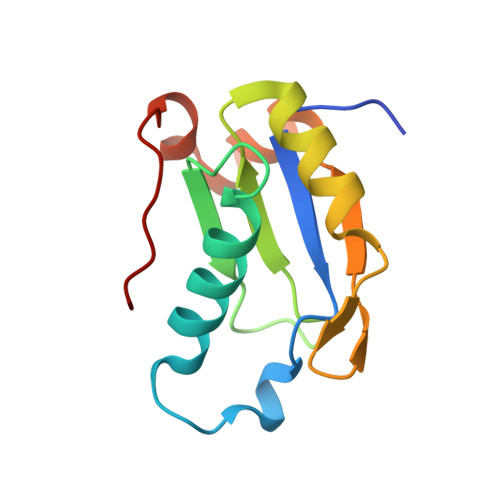
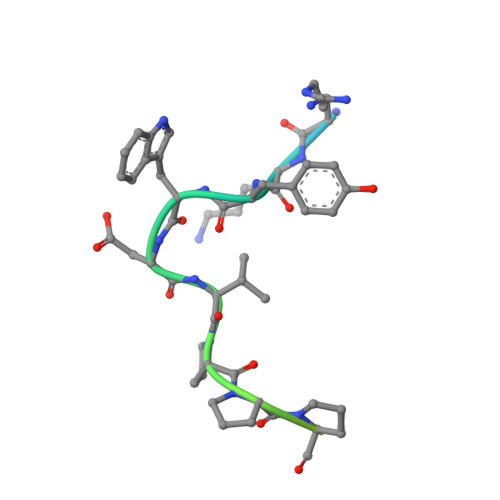
UHM-ULM interactions in the RBM39-U2AF65 splicing-factor complex.
Stepanyuk, G.A., Serrano, P., Peralta, E., Farr, C.L., Axelrod, H.L., Geralt, M., Das, D., Chiu, H.J., Jaroszewski, L., Deacon, A.M., Lesley, S.A., Elsliger, M.A., Godzik, A., Wilson, I.A., Wuthrich, K., Salomon, D.R., Williamson, J.R.(2016) Acta Crystallogr D Struct Biol 72: 497-511
- PubMed: 27050129
- DOI: https://doi.org/10.1107/S2059798316001248
- Primary Citation of Related Structures:
5CXT - PubMed Abstract:
RNA-binding protein 39 (RBM39) is a splicing factor and a transcriptional co-activator of estrogen receptors and Jun/AP-1, and its function has been associated with malignant progression in a number of cancers. The C-terminal RRM domain of RBM39 belongs to the U2AF homology motif family (UHM), which mediate protein-protein interactions through a short tryptophan-containing peptide known as the UHM-ligand motif (ULM). Here, crystal and solution NMR structures of the RBM39-UHM domain, and the crystal structure of its complex with U2AF65-ULM, are reported. The RBM39-U2AF65 interaction was confirmed by co-immunoprecipitation from human cell extracts, by isothermal titration calorimetry and by NMR chemical shift perturbation experiments with the purified proteins. When compared with related complexes, such as U2AF35-U2AF65 and RBM39-SF3b155, the RBM39-UHM-U2AF65-ULM complex reveals both common and discriminating recognition elements in the UHM-ULM binding interface, providing a rationale for the known specificity of UHM-ULM interactions. This study therefore establishes a structural basis for specific UHM-ULM interactions by splicing factors such as U2AF35, U2AF65, RBM39 and SF3b155, and a platform for continued studies of intermolecular interactions governing disease-related alternative splicing in eukaryotic cells.
Organizational Affiliation:
Department of Integrative Structural and Computational Biology, The Scripps Research Institute, La Jolla, CA 92037, USA.