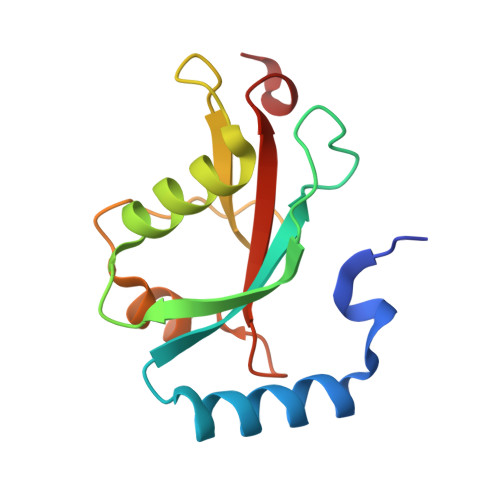
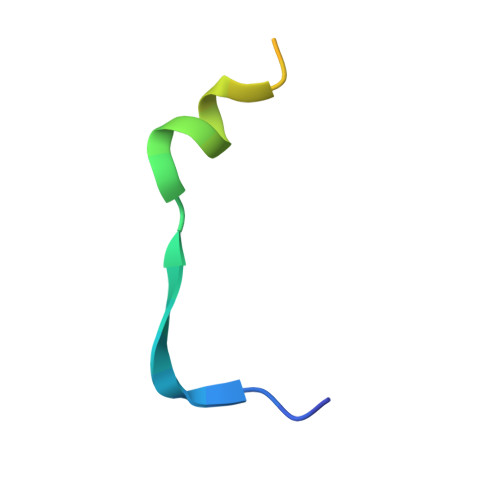
Structural basis of FYCO1 and MAP1LC3A interaction reveals a novel binding mode for Atg8-family proteins.
Cheng, X., Wang, Y., Gong, Y., Li, F., Guo, Y., Hu, S., Liu, J., Pan, L.(2016) Autophagy 12: 1330-1339
- PubMed: 27246247
- DOI: https://doi.org/10.1080/15548627.2016.1185590
- Primary Citation of Related Structures:
5CX3 - PubMed Abstract:
FYCO1 (FYVE and coiled-coil domain containing 1) functions as an autophagy adaptor in directly linking autophagosomes with the microtubule-based kinesin motor, and plays an essential role in the microtubule plus end-directed transport of autophagic vesicles. The specific association of FYCO1 with autophagosomes is mediated by its interaction with Atg8-family proteins decorated on the outer surface of autophagosome. However, the mechanistic basis governing the interaction between FYCO1 and Atg8-family proteins is largely unknown. Here, using biochemical and structural analyses, we demonstrated that FYCO1 contains a unique LC3-interacting region (LIR), which discriminately binds to mammalian Atg8 orthologs and preferentially binds to the MAP1LC3A and MAP1LC3B. In addition to uncovering the detailed molecular mechanism underlying the FYCO1 LIR and MAP1LC3A interaction, the determined FYCO1-LIR-MAP1LC3A complex structure also reveals a unique LIR binding mode for Atg8-family proteins, and demonstrates, first, the functional relevance of adjacent sequences C-terminal to the LIR core motif for binding to Atg8-family proteins. Taken together, our findings not only provide new mechanistic insight into FYCO1-mediated transport of autophagosomes, but also expand our understanding of the interaction modes between LIR motifs and Atg8-family proteins in general.
Organizational Affiliation:
a State Key Laboratory of Biorganic and Natural Products Chemistry , Shanghai , China.