Crystal structure of Trypanosoma cruzi protein in complex with ligand
Yang, Y.Y., Ko, T.P., Zheng, Y.Y., Liu, W.D., Chen, C.C., Guo, R.T.(2016) ACS Chem Biol
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
(2016) ACS Chem Biol
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Acidocalcisomal pyrophosphatase | 414 | Trypanosoma brucei brucei TREU927 | Mutation(s): 1 Gene Names: Tb11.02.4930 | 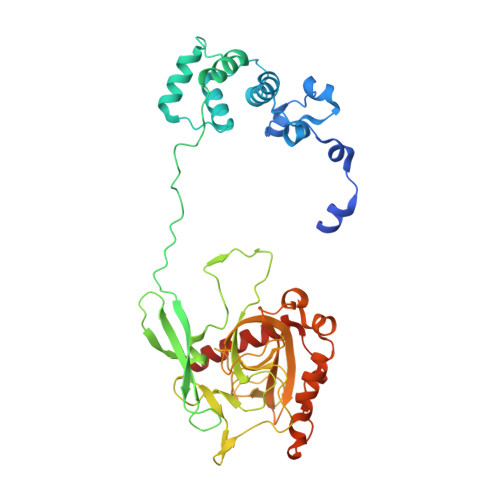 | |
UniProt | |||||
Find proteins for Q384W3 (Trypanosoma brucei brucei (strain 927/4 GUTat10.1)) Explore Q384W3 Go to UniProtKB: Q384W3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q384W3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| CIT Query on CIT | E [auth A], G [auth B], I [auth C], K [auth D] | CITRIC ACID C6 H8 O7 KRKNYBCHXYNGOX-UHFFFAOYSA-N |  | ||
| MG Query on MG | F [auth A], H [auth B], J [auth C], L [auth D] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 199.917 | α = 90 |
| b = 70.137 | β = 106.38 |
| c = 141.759 | γ = 90 |
| Software Name | Purpose |
|---|---|
| HKL-2000 | data scaling |
| CNS | refinement |
| PDB_EXTRACT | data extraction |
| HKL-2000 | data reduction |
| PHASER | phasing |