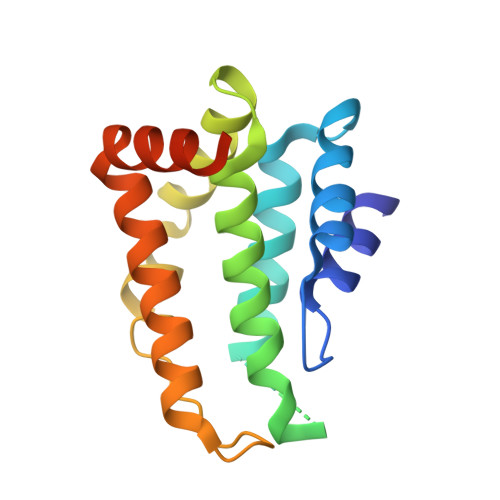
Structural and functional insight into the N-terminal domain of the clathrin adaptor Ent5 from Saccharomyces cerevisiae
Zhang, F., Song, Y., Ebrahimi, M., Niu, L., Teng, M.K., Li, X.(2016) Biochem Biophys Res Commun 477: 786-793
- PubMed: 27369074
- DOI: https://doi.org/10.1016/j.bbrc.2016.06.136
- Primary Citation of Related Structures:
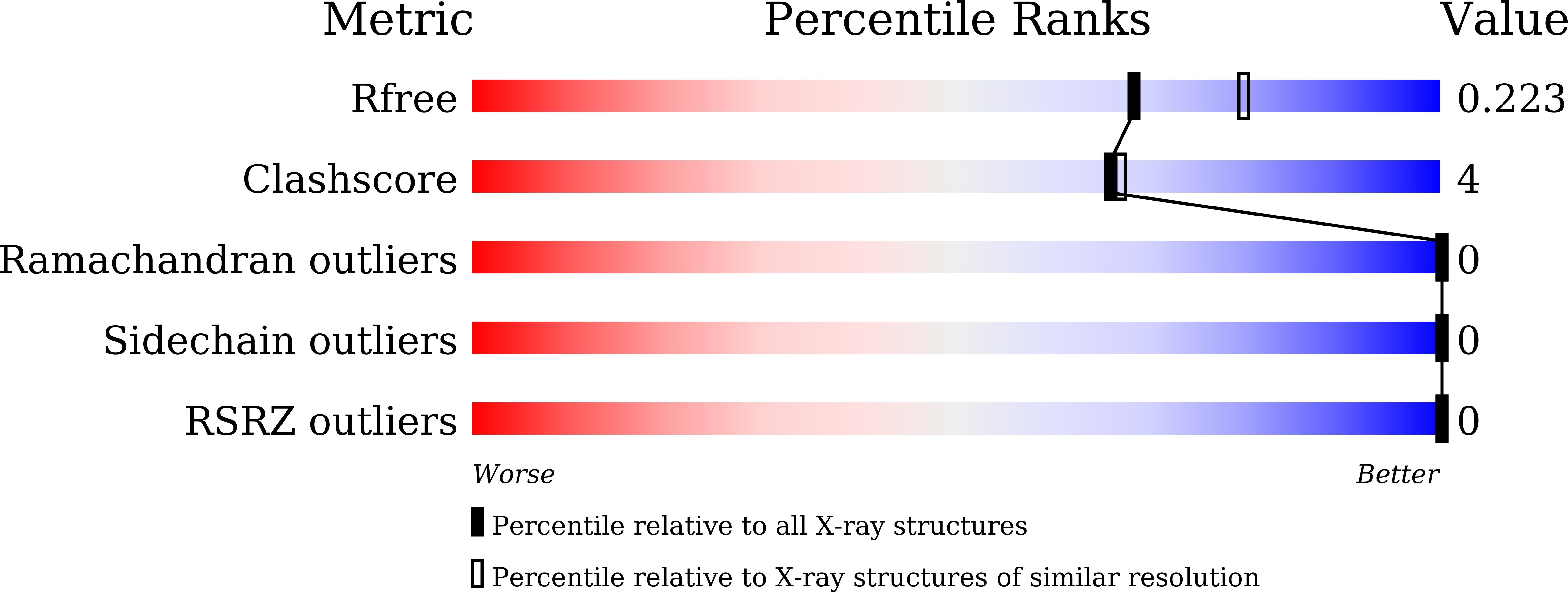
5CMW, 5CMY, 5J08 - PubMed Abstract:
Clathrin-coated vesicles (CCVs) play critical roles in multiple cellular processes, including nutrient uptake, endosome/lysosome biogenesis, pathogen invasion, regulation of signalling receptors, etc. Saccharomyces cerevisiae Ent5 (ScEnt5) is one of the two major adaptors supporting the CCV-mediated TGN/endosome traffic in yeast cells. However, the classification and phosphoinositide binding characteristic of ScEnt5 remain elusive. Here we report the crystal structures of the ScEnt5 N-terminal domain, and find that ScEnt5 contains an insertion α' helix that does not exist in other ENTH or ANTH domains. Furthermore, we investigate the classification of ScEnt5-N(31-191) by evolutionary history analyses and structure comparisons, and find that the ScEnt5 N-terminal domain shows different phosphoinositide binding property from rEpsin1 and rCALM. Above results facilitate the understanding of the ScEnt5-mediated vesicle coat formation process.
Organizational Affiliation:
Hefei National Laboratory for Physical Sciences at Microscale, Innovation Center for Cell Signaling Network, School of Life Science, 96 Jinzhai Road, Hefei, Anhui 230026, People's Republic of China; Key Laboratory of Structural Biology, Hefei Science Center of CAS, Chinese Academy of Sciences, 96 Jinzhai Road, Hefei, Anhui 230026, People's Republic of China.