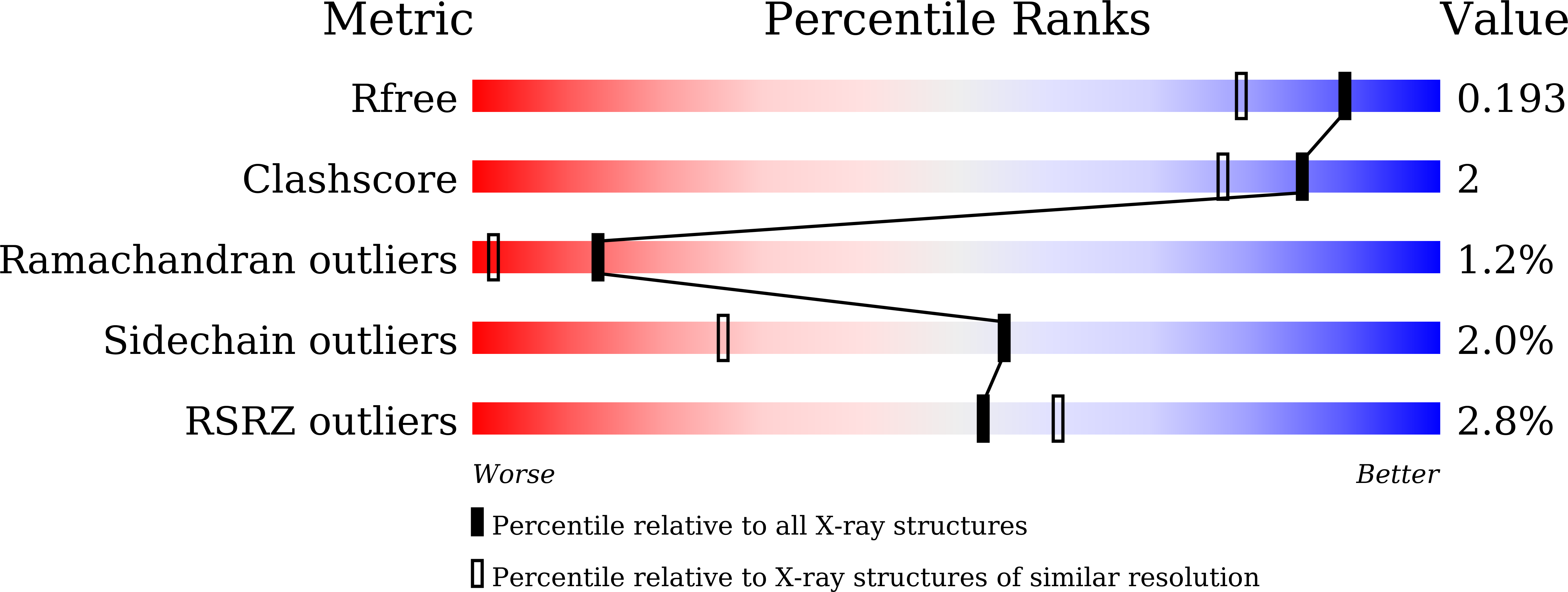
Biomineralization of a Cadmium Chloride Nanocrystal by a Designed Symmetrical Protein
Voet, A.R.D., Noguchi, H., Addy, C., Zhang, K.Y.J., Tame, J.R.H.(2015) Angew Chem Int Ed Engl 54: 9857-9860
- PubMed: 26136355
- DOI: https://doi.org/10.1002/anie.201503575
- Primary Citation of Related Structures:
5CHB - PubMed Abstract:
We have engineered a metal-binding site into the novel artificial β-propeller protein Pizza. This new Pizza variant carries two nearly identical domains per polypeptide chain, and forms a trimer with three-fold symmetry. The designed single metal ion binding site lies on the symmetry axis, bonding the trimer together. Two copies of the trimer associate in the presence of cadmium chloride in solution, and very high-resolution X-ray crystallographic analysis reveals a nanocrystal of cadmium chloride, sandwiched between two trimers of the protein. This nanocrystal, containing seven cadmium ions lying in a plane and twelve interspersed chloride ions, is the smallest reported to date. Our results indicate the feasibility of using rationally designed symmetrical proteins to biomineralize nanocrystals with useful properties.
Organizational Affiliation:
Structural Bioinformatics Team, Division of Structural and Synthetic Biology, Center for Life Science Technologies, RIKEN, 1-7-22, Suehiro, Tsurumi, Yokohama, 230-0045 (Japan). arnout.voet@riken.jp.