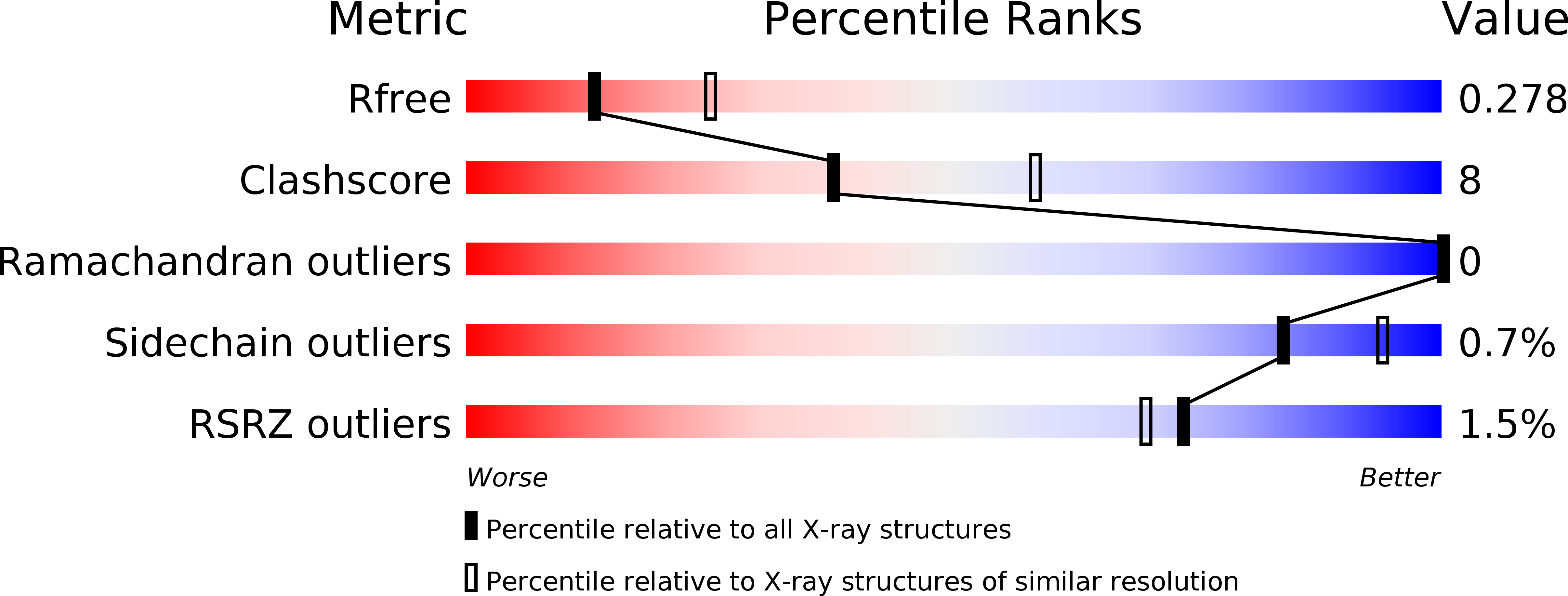
Structural and functional attributes of malaria parasite diadenosine tetraphosphate hydrolase
Sharma, A., Yogavel, M., Sharma, A.(2016) Sci Rep 6: 19981-19981
- PubMed: 26829485
- DOI: https://doi.org/10.1038/srep19981
- Primary Citation of Related Structures:
5CFI, 5CFJ - PubMed Abstract:
Malaria symptoms are driven by periodic multiplication cycles of Plasmodium parasites in human red blood corpuscles (RBCs). Malaria infection still accounts for ~600,000 annual deaths, and hence discovery of both new drug targets and drugs remains vital. In the present study, we have investigated the malaria parasite enzyme diadenosine tetraphosphate (Ap4A) hydrolase that regulates levels of signalling molecules like Ap4A by hydrolyzing them to ATP and AMP. We have tracked the spatial distribution of parasitic Ap4A hydrolase in infected RBCs, and reveal its unusual localization on the infected RBC membrane in subpopulation of infected cells. Interestingly, enzyme activity assays reveal an interaction between Ap4A hydrolase and the parasite growth inhibitor suramin. We also present a high resolution crystal structure of Ap4A hydrolase in apo- and sulphate- bound state, where the sulphate resides in the enzyme active site by mimicking the phosphate of substrates like Ap4A. The unexpected infected erythrocyte localization of the parasitic Ap4A hydrolase hints at a possible role of this enzyme in purinerigic signaling. In addition, atomic structure of Ap4A hydrolase provides insights for selective drug targeting.
Organizational Affiliation:
Structural and Computational Biology Group, International Centre for Genetic Engineering and Biotechnology (ICGEB), Aruna Asaf Ali Road, New Delhi, 110067, India.