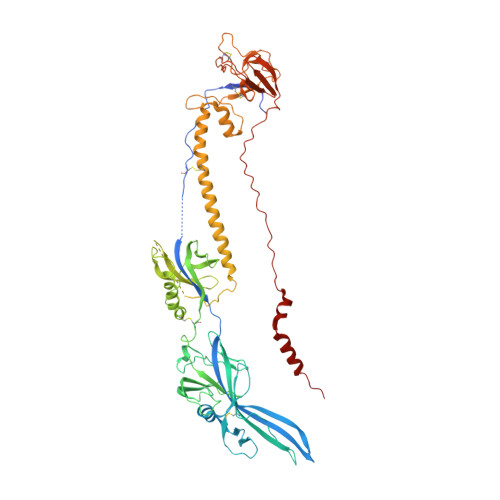
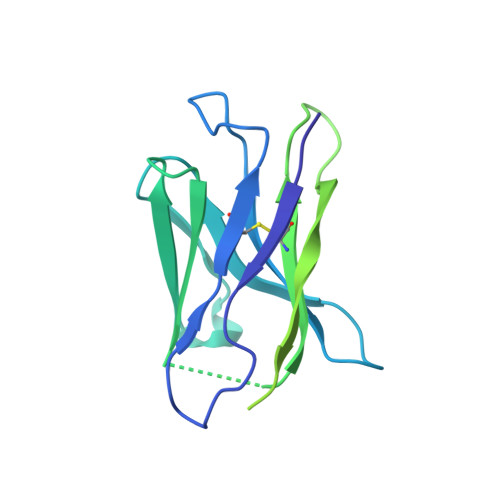
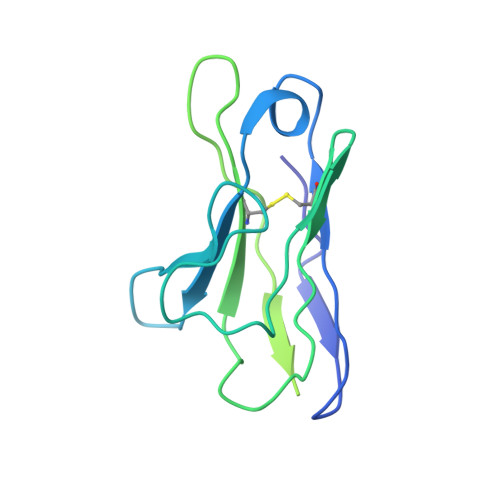
Structure of HCMV glycoprotein B in the postfusion conformation bound to a neutralizing human antibody.
Chandramouli, S., Ciferri, C., Nikitin, P.A., Calo, S., Gerrein, R., Balabanis, K., Monroe, J., Hebner, C., Lilja, A.E., Settembre, E.C., Carfi, A.(2015) Nat Commun 6: 8176-8176
- PubMed: 26365435
- DOI: https://doi.org/10.1038/ncomms9176
- Primary Citation of Related Structures:
5C6T - PubMed Abstract:
Human cytomegalovirus (HCMV) poses a significant threat to immunocompromised individuals and neonates infected in utero. Glycoprotein B (gB), the herpesvirus fusion protein, is a target for neutralizing antibodies and a vaccine candidate due to its indispensable role in infection. Here we show the crystal structure of the HCMV gB ectodomain bound to the Fab fragment of 1G2, a neutralizing human monoclonal antibody isolated from a seropositive subject. The gB/1G2 interaction is dominated by aromatic residues in the 1G2 heavy chain CDR3 protruding into a hydrophobic cleft in the gB antigenic domain 5 (AD-5). Structural analysis and comparison with HSV gB suggest the location of additional neutralizing antibody binding sites on HCMV gB. Finally, immunoprecipitation experiments reveal that 1G2 can bind to HCMV virion gB suggesting that its epitope is exposed and accessible on the virus surface. Our data will support the development of vaccines and therapeutic antibodies against HCMV infection.
Organizational Affiliation:
GSK Vaccines, 45 Sidney Street, Cambridge, Massachusetts 02139, USA.