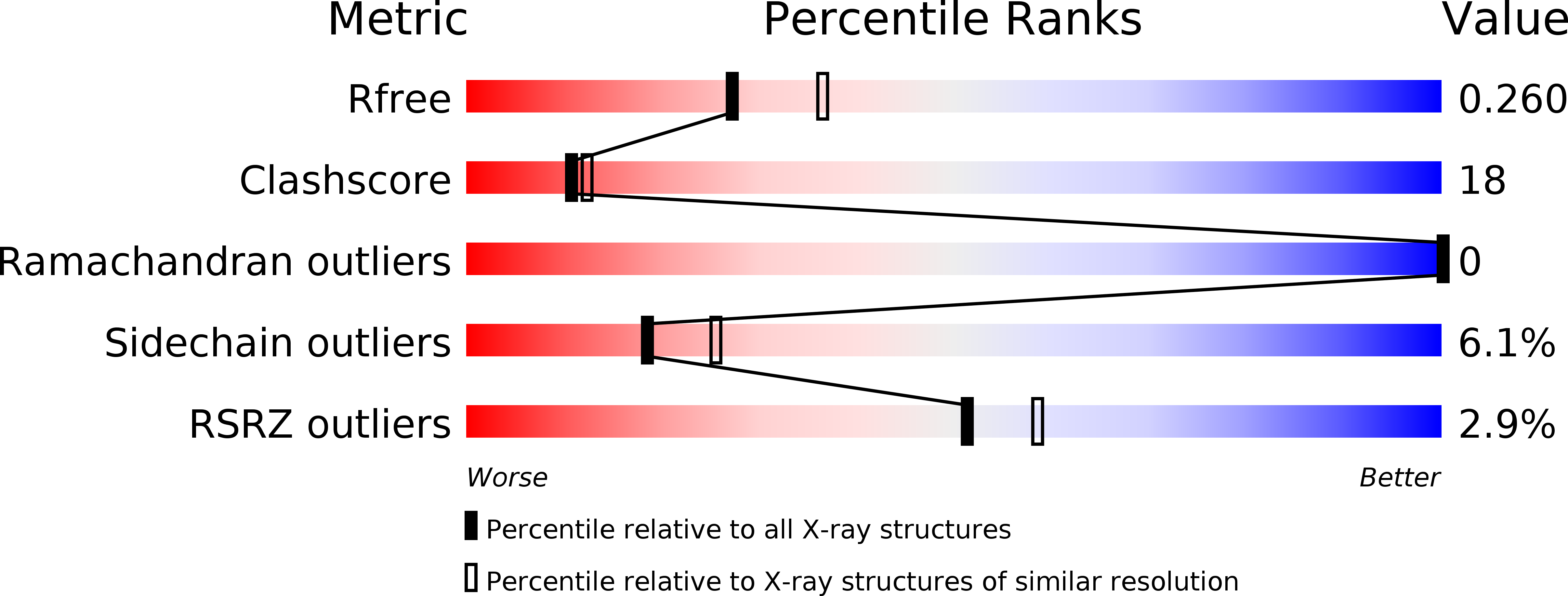
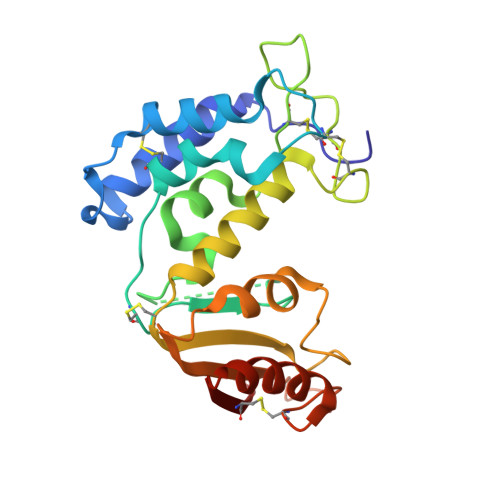
Porcine CD38 exhibits prominent secondary NAD(+) cyclase activity.
Ting, K.Y., Leung, C.F., Graeff, R.M., Lee, H.C., Hao, Q., Kotaka, M.(2016) Protein Sci 25: 650-661
- PubMed: 26660500
- DOI: https://doi.org/10.1002/pro.2859
- Primary Citation of Related Structures:
5BNF, 5BNI - PubMed Abstract:
Cyclic ADP-ribose (cADPR) mobilizes intracellular Ca(2+) stores and activates Ca(2+) influx to regulate a wide range of physiological processes. It is one of the products produced from the catalysis of NAD(+) by the multifunctional CD38/ADP-ribosyl cyclase superfamily. After elimination of the nicotinamide ring by the enzyme, the reaction intermediate of NAD(+) can either be hydrolyzed to form linear ADPR or cyclized to form cADPR. We have previously shown that human CD38 exhibits a higher preference towards the hydrolysis of NAD(+) to form linear ADPR while Aplysia ADP-ribosyl cyclase prefers cyclizing NAD(+) to form cADPR. In this study, we characterized the enzymatic properties of porcine CD38 and revealed that it has a prominent secondary NAD(+) cyclase activity producing cADPR. We also determined the X-ray crystallographic structures of porcine CD38 and were able to observe conformational flexibility at the base of the active site of the enzyme which allow the NAD(+) reaction intermediate to adopt conformations resulting in both hydrolysis and cyclization forming linear ADPR and cADPR respectively.
Organizational Affiliation:
School of Life Sciences, the Chinese University of Hong Kong, Hong Kong.