The postfusion structure of human-infecting Bourbon virus envelope glycoprotein implicates the host adaptation properties of thogotoviruses
Bai, C.Z., Qi, J.X., Wu, Y., Wang, X.J., Gao, G.F., Peng, R.C., Gao, F.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Envelope glycoprotein | 467 | Bourbon virus | Mutation(s): 0 | 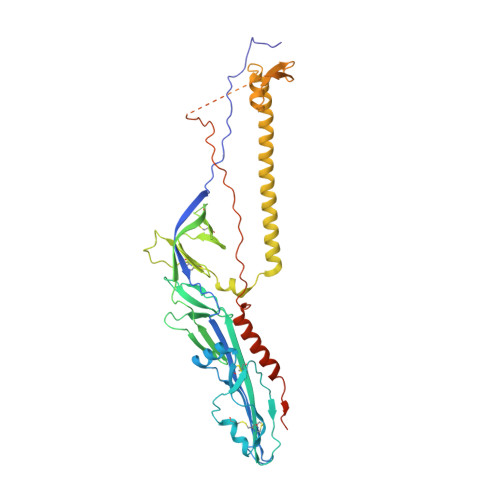 | |
UniProt | |||||
Find proteins for A0A140H4W8 (Bourbon virus) Explore A0A140H4W8 Go to UniProtKB: A0A140H4W8 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A140H4W8 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 102.255 | α = 90 |
| b = 102.255 | β = 90 |
| c = 134.472 | γ = 120 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Natural Science Foundation of China | China | 31741041 |
| National Natural Science Foundation of China | China | 81301465 |