Crystal structure of human Entervirus D68 RdRp in complex with NADPH
Wang, M.L., Zhang, Y., Dou, Y.S., Chen, Y.P., Lu, D.R., Jiang, H., Chen, Y.J., Li, L., Zhang, C.H., Shi, Q.L., Su, D.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
A newer entry is available that reflects an alternative modeling of the original data: 6L4R
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| RdRp | 457 | enterovirus D68 | Mutation(s): 0 | 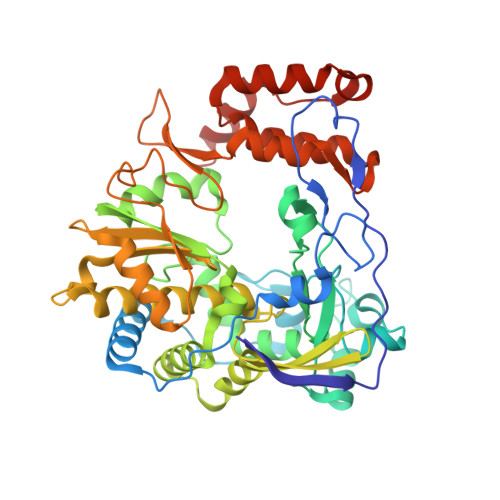 | |
UniProt | |||||
Find proteins for I6TFG7 (Human enterovirus D68) Explore I6TFG7 Go to UniProtKB: I6TFG7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | I6TFG7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| PEG Query on PEG | E [auth A], F [auth A], G [auth A] | DI(HYDROXYETHYL)ETHER C4 H10 O3 MTHSVFCYNBDYFN-UHFFFAOYSA-N |  | ||
| GOL Query on GOL | B [auth A], C [auth A], D [auth A] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 59.835 | α = 90 |
| b = 59.835 | β = 90 |
| c = 358.923 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |
| PHASER | phasing |