Trimeric structure of Vibrio cholerae Heat Shock Protein 15 at 2.3 Angstrom resolution
Roy Chowdhury, S., Sen, U.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Heat shock protein 15 | 127 | Vibrio cholerae O395 | Mutation(s): 0 Gene Names: VC0395_A2308 | 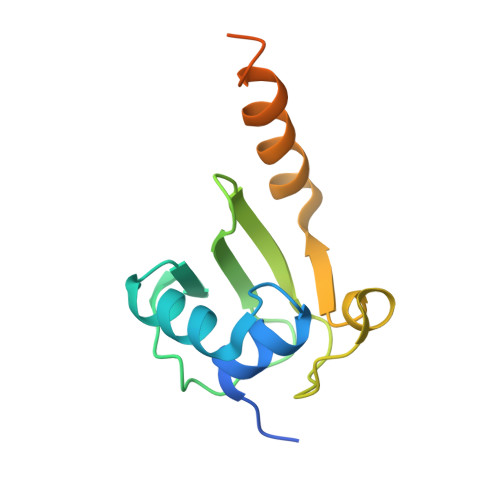 | |
UniProt | |||||
Find proteins for A0A0H3AM46 (Vibrio cholerae serotype O1 (strain ATCC 39541 / Classical Ogawa 395 / O395)) Explore A0A0H3AM46 Go to UniProtKB: A0A0H3AM46 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A0H3AM46 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| MPD Query on MPD | E [auth A], G [auth B], I [auth C] | (4S)-2-METHYL-2,4-PENTANEDIOL C6 H14 O2 SVTBMSDMJJWYQN-YFKPBYRVSA-N |  | ||
| SO4 Query on SO4 | D [auth A], F [auth B], H [auth C] | SULFATE ION O4 S QAOWNCQODCNURD-UHFFFAOYSA-L |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 53.65 | α = 90 |
| b = 90.28 | β = 103.92 |
| c = 81.09 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| iMOSFLM | data reduction |
| Aimless | data scaling |
| PHENIX | phasing |