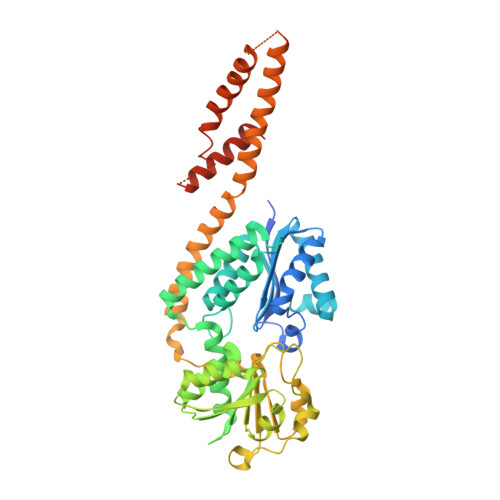
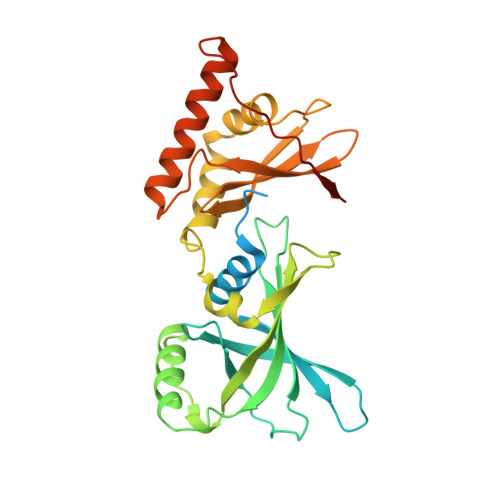
Crystal structure of Arabidopsis thaliana glutamyl-tRNAGlureductase in complex with NADPH and glutamyl-tRNAGlureductase binding protein
Zhao, A., Han, F.(2018) Photosynth Res 137: 443-452
- PubMed: 29785497
- DOI: https://doi.org/10.1007/s11120-018-0518-8
- Primary Citation of Related Structures:
5YJL - PubMed Abstract:
In higher plants, the tetrapyrrole biosynthesis pathway starts from the reaction catalyzed by the rate-limiting enzyme, glutamyl-tRNA Glu reductase (GTR). In Arabidopsis thaliana, GTR is controlled by post-transcriptional regulators such as GTR binding protein (GBP), which stimulates AtGTR activity. The NADPH-binding domain of AtGTR undergoes a substantial movement upon GBP binding. Here, we report the crystal structure of AtGTR-NADPH-GBP ternary complex. NADPH binding causes slight structural changes compared with the AtGTR-GBP binary complex, and possibly take a part of the space needed by the substrate glutamyl-tRNA Glu . The highly reactive sulfhydryl group of the active-site residue Cys144 shows an obvious rotation, which may facilitate the hydride transfer from NADPH to the thioester intermediate to form glutamate-1-semialdehyde. Furthermore, Lys271, Lys274, Ser275, Asn278, and Gln282 of GBP participate in the interaction between AtGTR and GBP, and the stimulating effect of GBP decreased when all of these residues were mutated to Ala. When the Cys144 of AtGTR was mutated to Ser, AtGTR activity could not be detected even in the presence of GBP.
Organizational Affiliation:
College of Forestry, Northwest A&F University, Yangling, 712100, Shaanxi, China.