Comparative structure analysis of the ETSi domain of ERG3 and its complex with the E74 promoter DNA sequence
Sharma, R., Gangwar, S.P., Saxena, A.K.(2018) Acta Crystallogr F Struct Biol Commun 74: 656-663
- PubMed: 30279318
- DOI: https://doi.org/10.1107/S2053230X1801110X
- Primary Citation of Related Structures:
5YBC, 5YBD - PubMed Abstract:
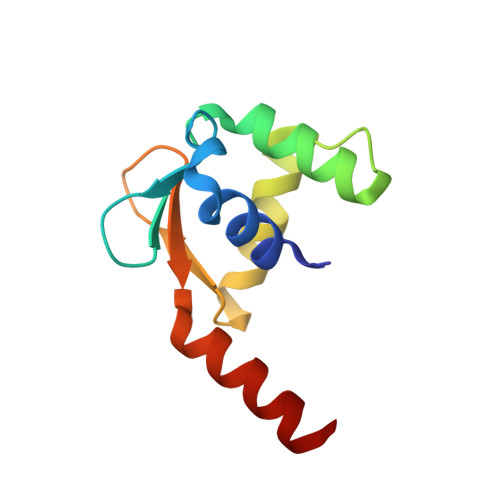
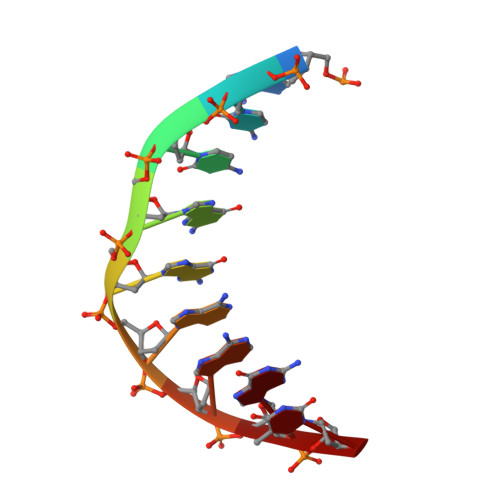
ERG3 (ETS-related gene) is a member of the ETS (erythroblast transformation-specific) family of transcription factors, which contain a highly conserved DNA-binding domain. The ETS family of transcription factors differ in their binding to promoter DNA sequences, and the mechanism of their DNA-sequence discrimination is little known. In the current study, crystals of the ETSi domain (the ETS domain of ERG3 containing a CID motif) in space group P4 1 2 1 2 and of its complex with the E74 DNA sequence (DNA 9 ) in space group C222 1 were obtained and their structures were determined. Comparative structure analysis of the ETSi domain and its complex with DNA 9 with previously determined structures of the ERGi domain (the ETS domain of ERG containing inhibitory motifs) in space group P6 5 2 1 2 and of the ERGi-DNA 12 complex in space group P4 1 2 1 2 were performed. The ETSi domain is observed as a homodimer in solution as well as in the crystallographic asymmetric unit. Superposition of the structure of the ETSi domain on that of the ERGi domain showed a major conformational change at the C-terminal DNA-binding autoinhibitory (CID) motif, while minor changes are observed in the loop regions of the ETSi-domain structure. The ETSi-DNA 9 complex in space group C222 1 forms a structure that is quite similar to that of the ERG-DNA 12 complex in space group P4 1 2 1 2. Upon superposition of the complexes, major conformational changes are observed at the 5' and 3' ends of DNA 9 , while the conformation of the core GGA nucleotides was quite conserved. Comparison of the ETSi-DNA 9 structure with known structures of ETS class 1 protein-DNA complexes shows the similarities and differences in the promoter DNA binding and specificity of the class 1 ETS proteins.
Organizational Affiliation:
Structural Biology Laboratory, School of Life Sciences, Jawaharlal Nehru University, New Delhi 110 067, India.