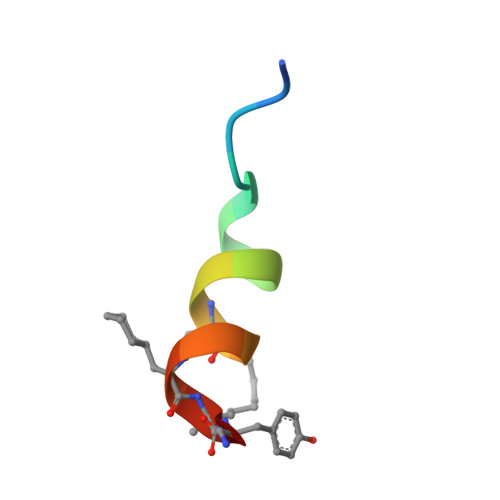
Targeted Inhibition of the NCOA1/STAT6 Protein-Protein Interaction
Lee, Y., Yoon, H., Hwang, S.M., Shin, M.K., Lee, J.H., Oh, M., Im, S.H., Song, J., Lim, H.S.(2017) J Am Chem Soc 139: 16056-16059
- PubMed: 29090910
- DOI: https://doi.org/10.1021/jacs.7b08972
- Primary Citation of Related Structures:
5Y7W - PubMed Abstract:
The complex formation between transcription factors (TFs) and coactivator proteins is required for transcriptional activity, and thus disruption of aberrantly activated TF/coactivator interactions could be an attractive therapeutic strategy. However, modulation of such protein-protein interactions (PPIs) has proven challenging. Here we report a cell-permeable, proteolytically stable, stapled helical peptide directly targeting nuclear receptor coactivator 1 (NCOA1), a coactivator required for the transcriptional activity of signal transducer and activator of transcription 6 (STAT6). We demonstrate that this stapled peptide disrupts the NCOA1/STAT6 complex, thereby repressing STAT6-mediated transcription. Furthermore, we solved the first crystal structure of a stapled peptide in complex with NCOA1. The stapled peptide therefore represents an invaluable chemical probe for understanding the precise role of the NCOA1/STAT6 interaction and an excellent starting point for the development of a novel class of therapeutic agents.
Organizational Affiliation:
Department of Chemistry and Division of Advanced Material Science, Pohang University of Science and Technology (POSTECH) , Pohang 37673, South Korea.