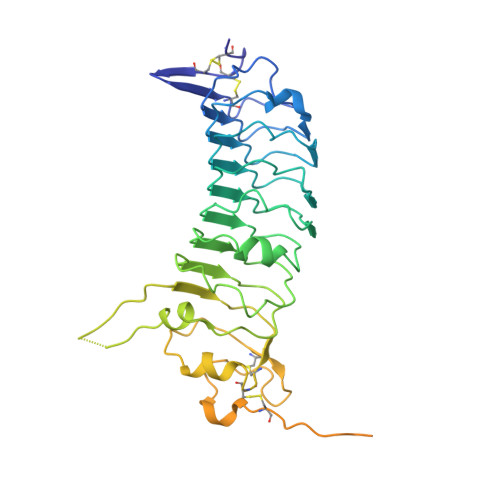
Structural basis of trans-synaptic interactions between PTP delta and SALMs for inducing synapse formation.
Goto-Ito, S., Yamagata, A., Sato, Y., Uemura, T., Shiroshima, T., Maeda, A., Imai, A., Mori, H., Yoshida, T., Fukai, S.(2018) Nat Commun 9: 269-269
- PubMed: 29348429
- DOI: https://doi.org/10.1038/s41467-017-02417-z
- Primary Citation of Related Structures:
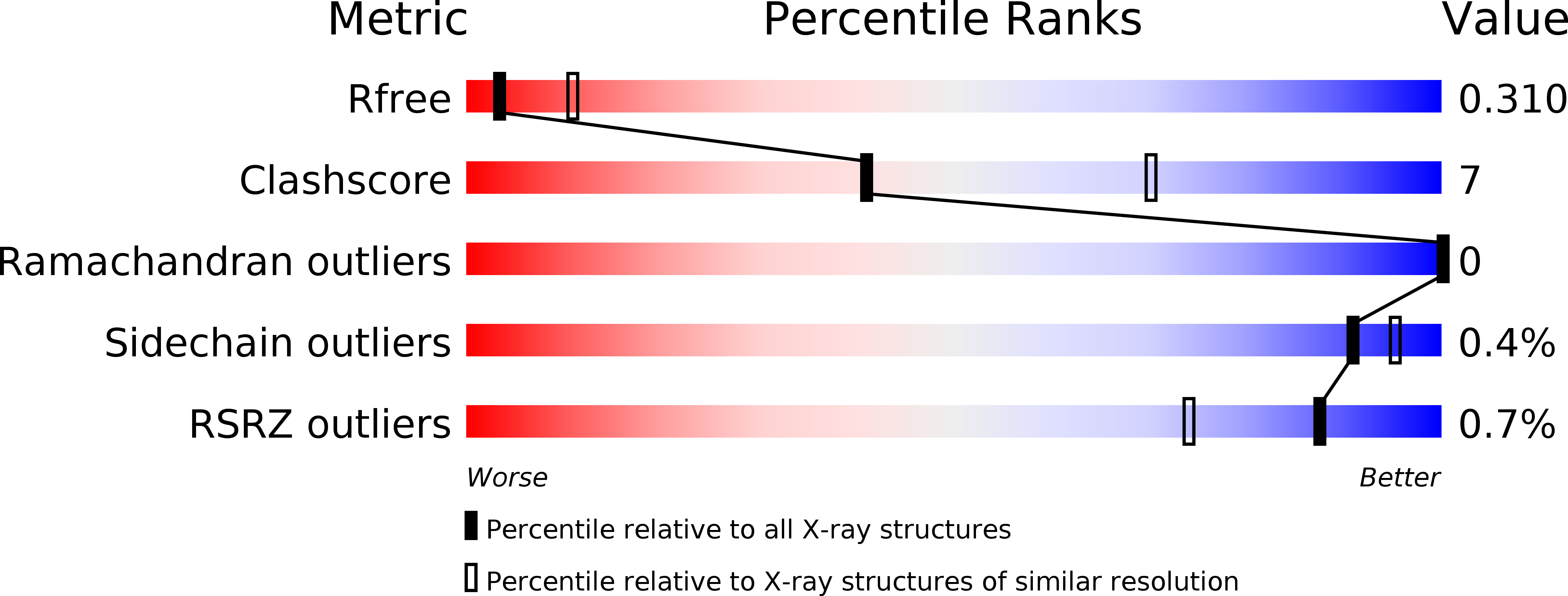
5XWS, 5XWT, 5XWU - PubMed Abstract:
Synapse formation is triggered by trans-synaptic interactions of cell adhesion molecules, termed synaptic organizers. Three members of type-II receptor protein tyrosine phosphatases (classified as type-IIa RPTPs; PTPδ, PTPσ and LAR) are known as presynaptic organizers. Synaptic adhesion-like molecules (SALMs) have recently emerged as a family of postsynaptic organizers. Although all five SALM isoforms can bind to the type-IIa RPTPs, only SALM3 and SALM5 reportedly have synaptogenic activities depending on their binding. Here, we report the crystal structures of apo-SALM5, and PTPδ-SALM2 and PTPδ-SALM5 complexes. The leucine-rich repeat (LRR) domains of SALMs interact with the second immunoglobulin-like (Ig) domain of PTPδ, whereas the Ig domains of SALMs interact with both the second and third Ig domains of PTPδ. Unexpectedly, the structures exhibit the LRR-mediated 2:2 complex. Our synaptogenic co-culture assay using site-directed SALM5 mutants demonstrates that presynaptic differentiation induced by PTPδ-SALM5 requires the dimeric property of SALM5.
Organizational Affiliation:
Institute of Molecular and Cellular Biosciences, The University of Tokyo, Tokyo, 113-0032, Japan.