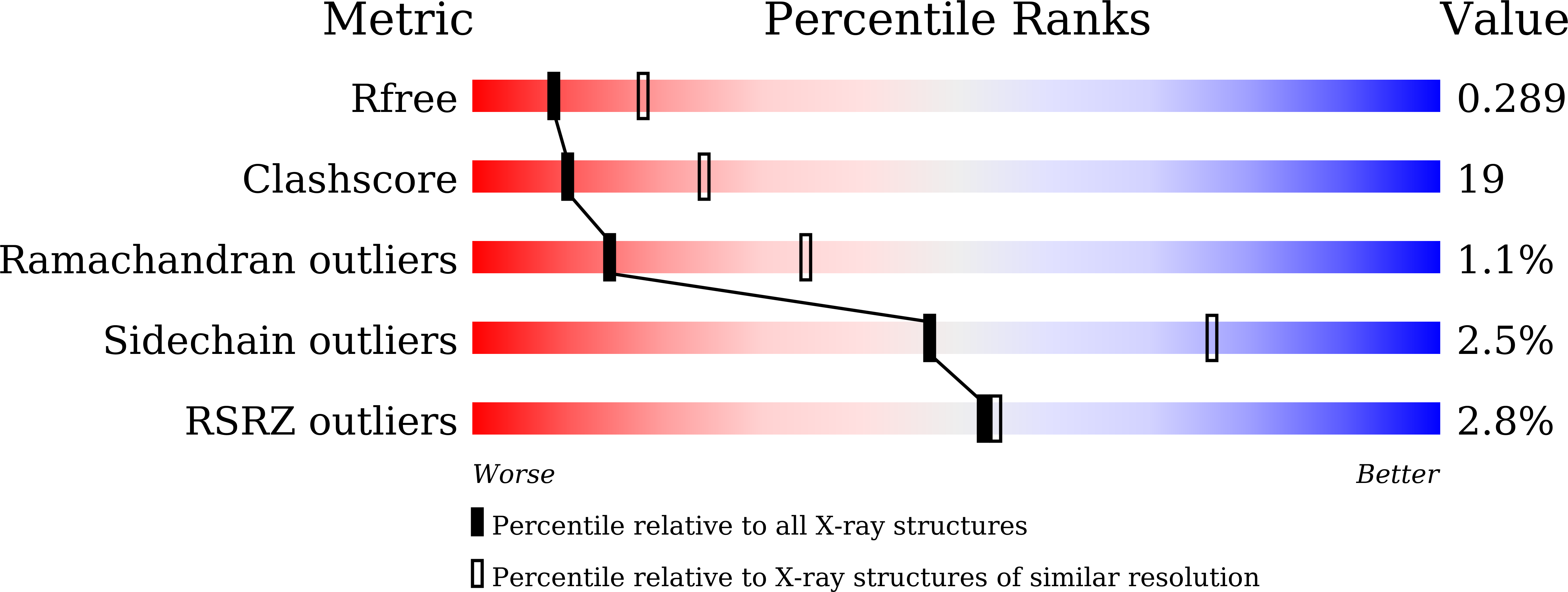
Crystal Structure of the Dimerized N Terminus of Porcine Circovirus Type 2 Replicase Protein Reveals a Novel Antiviral Interface
Luo, G., Zhu, X., Lv, Y., Lv, B., Fang, J., Cao, S., Chen, H., Peng, G., Song, Y.(2018) J Virol 92
- PubMed: 29976661
- DOI: https://doi.org/10.1128/JVI.00724-18
- Primary Citation of Related Structures:
5XOR - PubMed Abstract:
Two replicase (Rep) proteins, Rep and Rep', are encoded by porcine circovirus (PCV) ORF1; Rep is a full ORF1 transcript, and Rep' is a truncated transcript generated by splicing. These two proteins are crucial for the rolling-circle replication (RCR) of PCV. The N-terminal sequences of Rep and Rep' are identical and interact to form homo- or heterodimers. The three types of dimers perform different functions during replication. A structural examination of the interfacing termini has not been performed. In this study, a crystal structure of dimerized Rep protein N termini was resolved at 2.7 Å. The dimerized protein was maintained by nine intermolecular hydrogen bonds and 15 pairs of hydrophobic interactions. The amino acid residue Ile37 participates in 11 of the hydrophobic interactions, mostly with its side chain. To find the predominant sites for protein dimerization and virus replication, a series of mutant proteins and virus replicons were generated by alanine substitution. Of all the single amino acid substitutions, the mutation at Ile37 showed the greatest effect on protein dimerization and virus replication. A double mutation at Leu35 and Ile37 almost eliminated protein dimerization and had the greatest negative effect on virus replication. These studies demonstrate that Leu35 and Ile37 are the most important residues for protein dimerization and are crucial for virus replication. Our results also show that PCV replication can be decreased by disrupting the dimerization of Rep or Rep' at the N terminus, suggesting that the structural interface responsible for dimerization offers a promising antiviral target. IMPORTANCE Porcine circovirus type 2 (PCV2) is one of the most economically damaging pathogens affecting the swine industry. Although vaccines have been available for more than 10 years, the virus still remains prevalent. More effective strategies for disease prevention are clearly required. The Rep and Rep' proteins of the virus have identical N-terminal regions that interact with each other, allowing the formation of homo- or heterodimers. The heterodimer has crucial functions during different stages of viral replication. Here, we resolved the crystal structure of the Rep (Rep') dimerization domain. The individual residues involved in the intermolecular interaction were visualized in the protein structure, and several interactions were verified by mutant analysis. Our studies show that disrupting the interaction decreases viral replication, thus revealing a new target for the design of antiviral agents.
Organizational Affiliation:
State Key Laboratory of Agricultural Microbiology, Huazhong Agricultural University, Wuhan, China.