Insight into the transition between the open and closed conformations of Thermus thermophilus carboxypeptidase.
Okai, M., Yamamura, A., Hayakawa, K., Tsutsui, S., Miyazono, K.I., Lee, W.C., Nagata, K., Inoue, Y., Tanokura, M.(2017) Biochem Biophys Res Commun 484: 787-793
- PubMed: 28161633
- DOI: https://doi.org/10.1016/j.bbrc.2017.01.167
- Primary Citation of Related Structures:
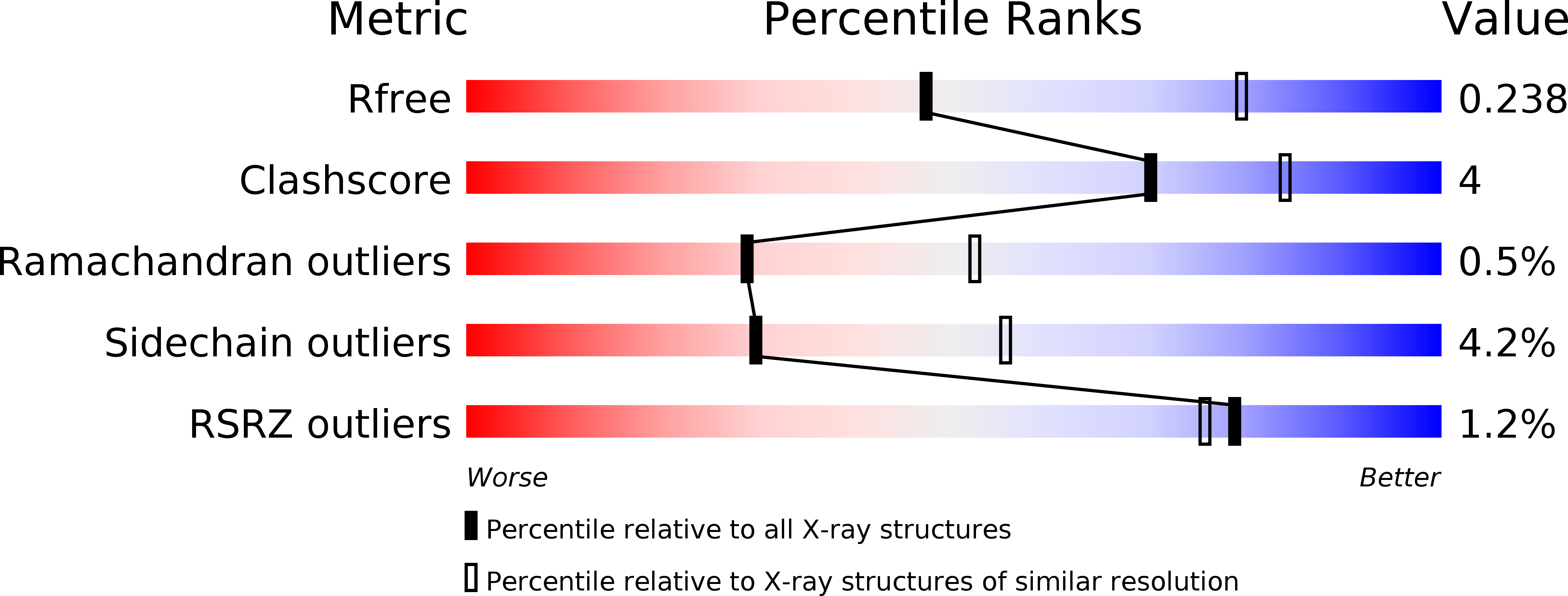
5WVU - PubMed Abstract:
Carboxypeptidase cleaves the C-terminal amino acid residue from proteins and peptides. Here, we report the functional and structural characterizations of carboxypeptidase belonging to the M32 family from the thermophilic bacterium Thermus thermophilus HB8 (TthCP). TthCP exhibits a relatively broad specificity for both hydrophilic (neutral and basic) and hydrophobic (aliphatic and aromatic) residues at the C-terminus and shows optimal activity in the temperature range of 75-80 °C and in the pH range of 6.8-7.2. Enzyme activity was significantly enhanced by cobalt or cadmium and was moderately inhibited by Tris at 25 °C. We also determined the crystal structure of TthCP at 2.6 Å resolution. Two dimer types of TthCP are present in the crystal. One type consists of two subunits in different states, open and closed, with a C α RMSD value of 2.2 Å; the other type consists of two subunits in the same open state. This structure enables us to compare the open and closed states of an M32 carboxypeptidase. The TthCP subunit can be divided into two domains, L and S, which are separated by a substrate-binding groove. The L and S domains in the open state are almost identical to those in the closed state, with C α RMSD values of 0.84 and 0.53 Å, respectively, suggesting that the transition between the open and closed states proceeds with a large hinge-bending motion. The superimposition between the closed states of TthCP and BsuCP, another M32 family member, revealed that most putative substrate-binding residues in the grooves are oriented in the same direction.
Organizational Affiliation:
Department of Applied Biological Chemistry, Graduate School of Agricultural and Life Sciences, The University of Tokyo, 1-1-1 Yayoi, Bunkyo-ku, Tokyo 113-8657, Japan; Tokyo University of Marine Science and Technology, 4-5-7 Konan, Minato-ku, Tokyo 108-8477, Japan.