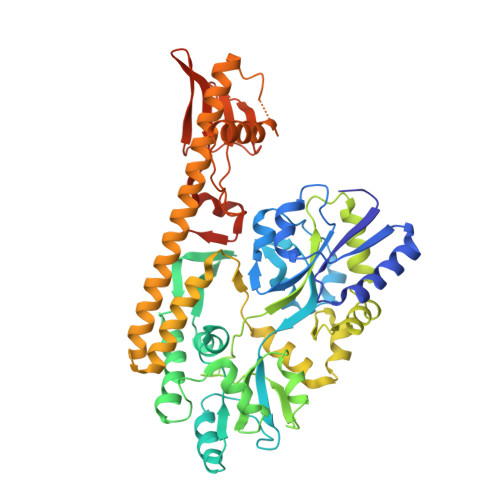
Crystal structure of the sensor domain of BaeS from Serratia marcescens FS14
Zhang, Y., Qiu, S., Jia, S., Xu, D., Ran, T., Wang, W.(2017) Proteins 85: 1784-1790
- PubMed: 28544098
- DOI: https://doi.org/10.1002/prot.25326
- Primary Citation of Related Structures:
5WVM, 5WVN - PubMed Abstract:
The sensor histidine kinases of two-component signal-transduction systems (TCSs) are essential for bacteria to adapt to variable environmental conditions. The two-component regulatory system BaeS/R increases multidrug and metal resistance in Salmonella and Escherichia coli. In this study, we report the X-ray structure of the periplasmic sensor domain of BaeS from Serratia marcescens FS14. The BaeS sensor domain (34-160) adopts a mixed α/β-fold containing a central four-stranded antiparallel β-sheet flanked by a long N-terminal α-helix and additional loops and a short C-terminal α-helix on each side. Structural comparisons revealed that it belongs to the PDC family with a remarkable difference in the orientation of the helix α2. In the BaeS sensor domain, this helix is situated perpendicular to the long helix α1 and holds helix α1 in the middle with the beta sheet, whereas in other PDC domains, helix α2 is parallel to helix α1. Because the helices α1 and α2 is involved in the dimeric interface, this difference implies that BaeS uses a different dimeric interface compared with other PDC domains. Proteins 2017; 85:1784-1790. © 2017 Wiley Periodicals, Inc.
Organizational Affiliation:
Key Laboratory of Agricultural and Environmental Microbiology, Ministry of Agriculture, College of Life Sciences, Department of Microbiology, Nanjing Agricultural University, Nanjing, 210095, China.