Crystal Structures of Anti-apoptotic BFL-1 and Its Complex with a Covalent Stapled Peptide Inhibitor.
Harvey, E.P., Seo, H.S., Guerra, R.M., Bird, G.H., Dhe-Paganon, S., Walensky, L.D.(2018) Structure 26: 153-160.e4
- PubMed: 29276033
- DOI: https://doi.org/10.1016/j.str.2017.11.016
- Primary Citation of Related Structures:
5WHH, 5WHI - PubMed Abstract:
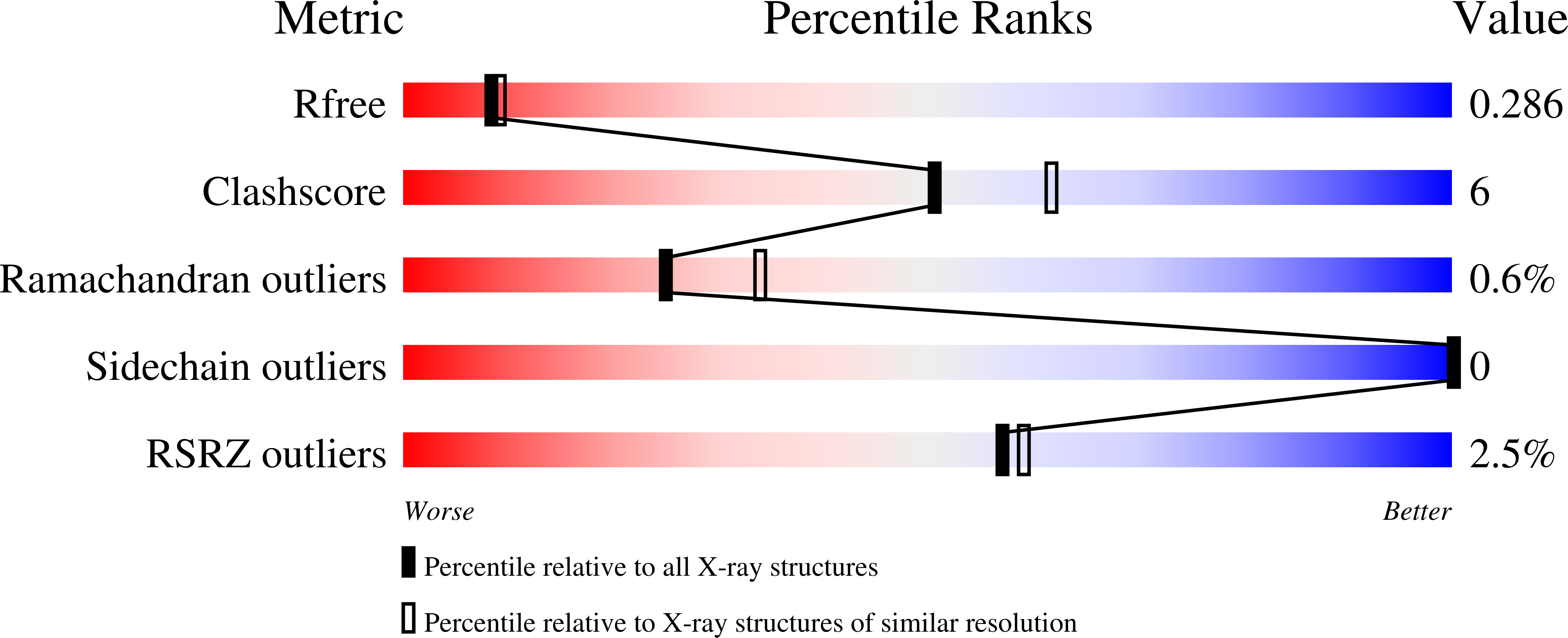
BCL-2 family proteins are high-priority cancer targets whose structures provide essential blueprints for drug design. Whereas numerous structures of anti-apoptotic BCL-2 protein complexes with α-helical BH3 peptides have been reported, the corresponding panel of apo structures remains incomplete. Here, we report the crystal structure of apo BFL-1 at 1.69-Å resolution, revealing similarities and key differences among unliganded anti-apoptotic proteins. Unlike all other BCL-2 proteins, apo BFL-1 contains a surface-accessible cysteine within its BH3-binding groove, allowing for selective covalent targeting by a NOXA BH3-based stapled peptide inhibitor. The crystal structure of this complex demonstrated the sulfhydryl bond and fortuitous interactions between the acrylamide-bearing moiety and a newly formed hydrophobic cavity. Comparison of the apo and BH3-liganded structures further revealed an induced conformational change. The two BFL-1 structures expand our understanding of the surface landscapes available for therapeutic targeting so that the apoptotic blockades of BFL-1-dependent cancers can be overcome.
Organizational Affiliation:
Department of Pediatric Oncology, Dana-Farber Cancer Institute, Boston, MA 02215, USA; Linde Program in Cancer Chemical Biology, Dana-Farber Cancer Institute, Boston, MA 02215, USA.