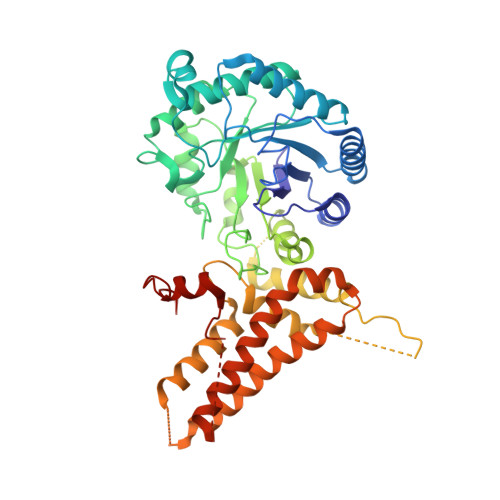
Structural insights into the substrate binding adaptability and specificity of human O-GlcNAcase.
Li, B., Li, H., Hu, C.W., Jiang, J.(2017) Nat Commun 8: 666-666
- PubMed: 28939839
- DOI: https://doi.org/10.1038/s41467-017-00865-1
- Primary Citation of Related Structures:
5VVO, 5VVT, 5VVU, 5VVV, 5VVX - PubMed Abstract:
The O-linked β-N-acetyl glucosamine (O-GlcNAc) modification dynamically regulates the functions of numerous proteins. A single human enzyme O-linked β-N-acetyl glucosaminase (O-GlcNAcase or OGA) hydrolyzes this modification. To date, it remains largely unknown how OGA recognizes various substrates. Here we report the structures of OGA in complex with each of four distinct glycopeptide substrates that contain a single O-GlcNAc modification on a serine or threonine residue. Intriguingly, these glycopeptides bind in a bidirectional yet conserved conformation within the substrate-binding cleft of OGA. This study provides fundamental insights into a general principle that confers the substrate binding adaptability and specificity to OGA in O-GlcNAc regulation.O-linked β-N-acetyl glucosamine (O-GlcNAc) is an important protein modification that is hydrolyzed by O-GlcNAcase (OGA). Here the authors give insights into OGA substrate recognition by presenting four human OGA structures complexed with glycopeptide substrates containing a single O-GlcNAc modification on either a serine or threonine.
Organizational Affiliation:
Pharmaceutical Sciences Division, School of Pharmacy, University of Wisconsin-Madison, Madison, WI, 53705, USA.