The structure of PilA fromAcinetobacter baumanniiAB5075 suggests a mechanism for functional specialization inAcinetobactertype IV pili.
Ronish, L.A., Lillehoj, E., Fields, J.K., Sundberg, E.J., Piepenbrink, K.H.(2019) J Biol Chem 294: 218-230
- PubMed: 30413536
- DOI: https://doi.org/10.1074/jbc.RA118.005814
- Primary Citation of Related Structures:
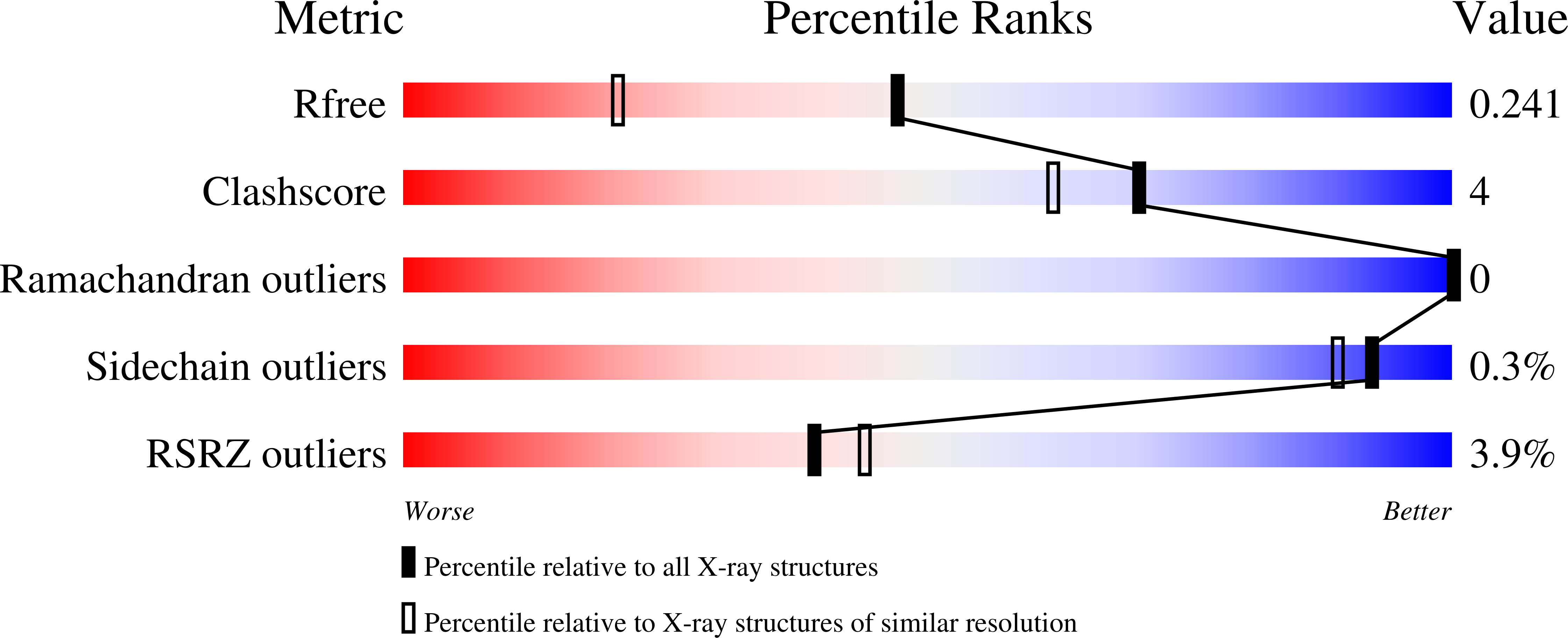
5VAW - PubMed Abstract:
Type IV pili (T4P) are bacterial appendages composed of protein subunits, called pilins, noncovalently assembled into helical fibers. T4P are essential, in many bacterial species, for processes as diverse as twitching motility, natural competence, biofilm or microcolony formation, and host cell adhesion. The genes encoding type IV pili are found universally in the Gram-negative, aerobic, nonflagellated, and pathogenic coccobacillus Acinetobacter baumannii , but there is considerable variation in PilA, the major protein subunit, both in amino acid sequence and in glycosylation patterns. Here we report the X-ray crystal structure of PilA from AB5075, a recently characterized, highly virulent isolate, at 1.9 Å resolution and compare it to homologues from A. baumannii strains ACICU and BIDMC57, which are C-terminally glycosylated. These structural comparisons revealed that PilA AB5075 exhibits a distinctly electronegative surface chemistry. To understand the functional consequences of this change in surface electrostatics, we complemented a Δ pilA knockout strain with divergent pilA genes from ACICU, BIDMC57, and AB5075. The resulting transgenic strains showed differential twitching motility and biofilm formation while maintaining the ability to adhere to epithelial cells. PilA AB5075 and PilA ACICU , although structurally similar, promote different characteristics, favoring twitching motility and biofilm formation, respectively. These results support a model in which differences in pilus electrostatics affect the equilibrium of microcolony formation, which in turn alters the balance between motility and biofilm formation in Acinetobacter .
Organizational Affiliation:
Department of Biochemistry, University of Nebraska-Lincoln, Lincoln, Nebraska 68588.